Information Technology Reference
In-Depth Information
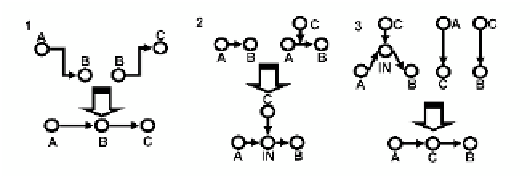
Fig. 4.6. Inference rules used in assembling component-to-component and component-to-pathway
relationships in (Mendoza
et al.
1998; Mendoza
et al.
1999). These rules incorporate node to
pathway relationships by introducing unknown intermediary nodes, then collapse the intermediary
nodes with known nodes in the system if additional information is available. Figure adapted from
(Li
et al
. 2006).
Experimental data is also used to design the Boolean/ transition rules of the
nodes. The operator NOT is used to model an inhibition interaction between two
components. The operator OR is used when two positive regulators can
independently activate a target node whereas the operator AND is used when
both nodes are required for the activation of the target node. Various types of
interactions and regulatory relationships can be described by using these three
operators. Below we will review examples of Boolean models used to simulate
the dynamics of various biological systems.
Plant Biology:
Signaling pathways and gene regulatory networks in plants are
interesting examples of the complex regulation of genes during morphogenesis
and in response to environmental signals. The development of a complex
structure such as a flower has been successfully described with Boolean models.
Network assembly and a Boolean model of the genes involved in
Arabidopsis
thaliana
flower morphogenesis indicated six attractors, four of which correspond
to the observed patterns of gene expression in four floral parts, petals, sepals,
stamens and carpels (Mendoza
et al
. 1998). The fifth attractor corresponds to the
cells that are not competent to flower and the sixth attractor, though not found in
wild-type plants, can be induced experimentally. The model incorporates relative
interaction weights and activation thresholds in addition to the Boolean rules
describing regulation of each node, so that a gene is active if the weighted input
of all the genes regulating it exceeds its activation threshold. A follow up study
of this network identified the regulatory constraints and functional feedback
circuits that lead to stable gene expression patterns (Mendoza
et al
. 1999).
An extension (Espinosa-Soto
et al
. 2004) of the network published by (Li
et al.
2006) implemented two expression levels for eight nodes and three for seven
nodes. Simulations with all possible initial conditions
converged to a few steady
gene activity states that match gene
expression profiles observed experimentally