Biomedical Engineering Reference
In-Depth Information
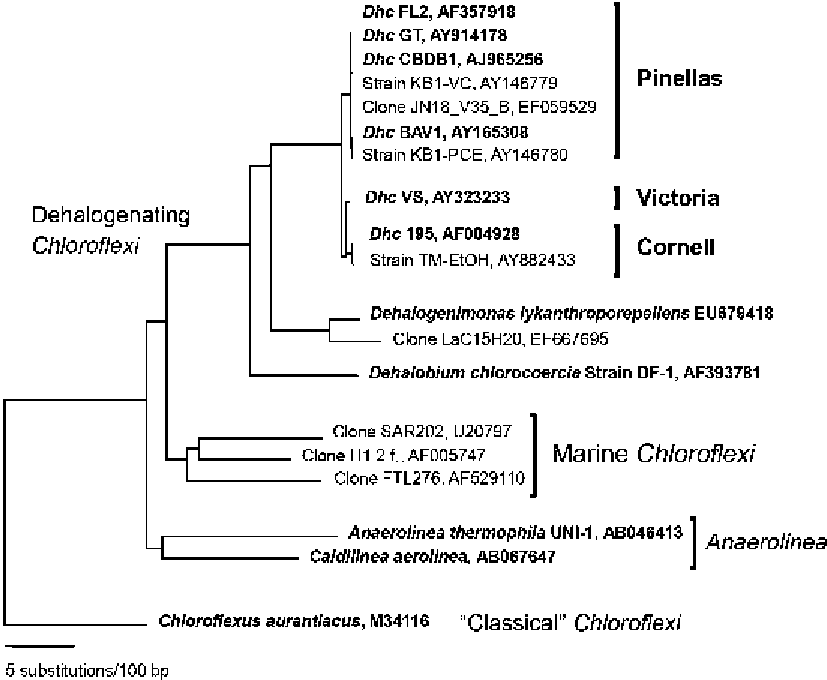
Figure 2.3. Phylogenetic affiliation of Dhc and related dechlorinators in the phylum Chloroflexi
(green non-sulfur bacteria). Represented are the two main branches, the Thermomicrobia/Chloro-
flexus branch (represented by Chloroflexus aurantiacus) and the branch containing Dhc and
relatives. The Neighbor-Joining tree was generated using an alignment of 1,350 bases of the 16S
rRNA gene of the Chloroflexi, with focus on Dehalococcoides relatives. At the top is the Dehalo-
coccoides cluster, followed by 16S rRNA gene sequences of dechlorinating Chloroflexi isolates
that are not detected with the Dehalococcoides targeted PCR assays. The Anaerolinea and marine
clusters are represented by isolates and environmental clone sequences, respectively. Pure
cultures are indicated by bold font. “Classical” Chloroflexi isolates form deeper branches, and
are in the lineage with Chloroflexus aurantiacus. The tree is based on a matrix calculated with
maximum likelihood analysis with Juke's-Cantor adjustments. Scale bar represents 5% sequence
divergence.
subgroup, 32 with the Cornell subgroup, and 8 with the Victoria subgroup. While this survey is
not statistically valid, it suggests that members of the Pinellas subgroup are most commonly
encountered in contaminated environments (where most of the sequences were retrieved).
It should be noted that this grouping has no bearing on dechlorination activity. Members of
all three groups dechlorinate chlorinated ethenes and strain VS (Victoria subgroup) and strains
BAV1 and GT (Pinellas subgroup) grow with
cis
-DCE and VC as electron acceptors. An
important observation is that some members of the Pinellas subgroup share identical 16S
Search WWH ::

Custom Search