Biomedical Engineering Reference
In-Depth Information
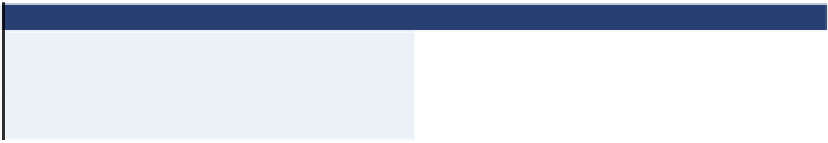
Table 6.1. Dehalococcoides qPCR Analysis Rules of Thumb
Dhc 16S rRNA (gene copies per L)
Interpretation
10
3
or lower
Suboptimal Dhc to sustain dechlorination rates
10
4
-10
6
May sustain appreciable dechlorination rates
10
7
or greater
Usually associated with high rates of dechlorination
and ethene production
There is some uncertainty regarding the numbers of
Dhc
that are needed for acceptable
rates of dechlorination, or that would indicate complete biodegradation to ethene is occurring.
However, some rules of thumb have been developed for interpreting
Dhc
results (Lebr ´ n et al.,
2011a
,
b
). In general,
Dhc
concentrations less than 10
3
per L indicate suboptimal concentrations
of
Dhc
cells in groundwater, and such levels may not be capable of sustaining complete
dechlorination (van der Zaan et al.,
2010
; Lebr ´ n et al.,
2011a
). Concentrations of 10
4
-10
6
per
L may provide acceptable dechlorination rates, while concentrations greater than 10
6
per L
often achieve high rates suitable for effective bioremediation (van der Zaan et al.,
2010
;
Lu et al.,
2006
,
2009
) and are considered the threshold concentration. These rules of thumb
for
Dhc
data interpretation also are shown in Table
6.1
.
As noted above, the limitations of qPCR must be considered when analyzing data obtained
from environmental samples. For example, even when sufficient numbers of
Dhc
cells are
present, complete reductive dechlorination may not occur, because the
Dhc
group contains a
diverse number of species and strains, with some capable of transforming TCE through to
ethene, and others capable of transformation only to DCE or VC. Research studies discovered a
correlation between particular genes (named “dehalogenases”) and the varied ability to trans-
form these compounds. One example is the
vcrA
gene mentioned in the introduction, which is
responsible for the final dehalogenation step reducing VC to ethene, and is not found in the
genomes of all known
Dhc
. Although qPCR analysis of the
Dhc
16S rRNA gene may indicate
greater than 10
8
cells per L (a quantity likely capable of sustained and vigorous reductive
dechlorination), if a sufficient quantity of these cells are
Dhc
species lacking the
vcrA
gene, VC
will accumulate. Thus, analysis using qPCR for determining the concentration of the
vcrA
gene
(and/or other VC reductase genes) may be required.
6.5.5 Conclusion
The development of qPCR as a viable molecular biological tool for environmental analyses
provides a tremendous opportunity for collecting data in support of bioaugmentation design,
implementation and performance monitoring. However, as with chemical data obtained from
the environment, careful planning and a consistent approach are critical to ensure an accurate
assessment of site conditions is made. This is especially true given the nature of qPCR, where
sampling variability may result in reported concentrations orders of magnitude different from
actual concentrations.
6.6 FLUORESCENT
IN SITU
HYBRIDIZATION
6.6.1 Introduction
Fluorescent
in situ
hybridization (FISH) is a molecular technique used to detect genes
within microbes of interest in environmental samples, including
Dhc
(Madigan et al.,
2003
;
Yang and Zeyer,
2003
). Through the use of fluorescently-labeled molecular probes, microbial






Search WWH ::

Custom Search