Biomedical Engineering Reference
In-Depth Information
Cl
Cl
Cl
Cl
Cl
H
H
H
H
H
2H
+
+ 2e
-
2H
+
+ 2e
-
2H
+
+ 2e
-
2H
+
+ 2e
-
C
C
C
C
C
C
C
C
C
C
H
+
+ Cl
-
H
+
+ Cl
-
H
+
+ Cl
-
H
+
+ Cl
-
Cl
Cl
Cl
H
Cl
H
Cl
H
H
H
pceA
tceA
Dhc
strain 195
tceA
Dhc
strain FL2
bvcA
Dhc
strain BAV1
?
vcrA
Dhc
strain VS,
Dhc
strain GT
Figure 2.7. Dhc RDase genes implicated in reductive dechlorination of chlorinated ethenes.
Although the RDase genes identified to date are only a subset of the total number of RDase
genes contributing to the reductive dechlorination of chlorinated ethenes, the quantitative assess-
ment of tceA, vcrA, bvcA and Dhc 16S rRNA genes has proven useful for prognostic site assess-
ment and bioremediation monitoring. Strains GT and VS dechlorinate TCE but do not possess tceA
and the gene(s) encoding this function may serve as an additional biomarker for this process. VcrA
has been biochemically characterized and dechlorinates all DCE isomers and VC in in vitro assays
(M ¨ ller et al.,
2004
; Rosner et al.,
1997
). Transcriptional analysis implicated BvcA in VC dechlori-
nation (Krajmalnik-Brown et al.,
2004
) but its involvement in DCE dechlorination has yet to be
demonstrated.
implicated in chlorinated ethene dechlorination has been identified (Ritalahti et al.,
2006
).
To achieve comprehensive monitoring of the numerous
Dhc
strains with distinct RDase
genes contributing to chlorinated ethene detoxification, function must be assigned to the
remaining identified RDase genes, and such efforts are underway in several laboratories. In
addition, other process-specific
Dhc
biomarker genes are being sought including hydrogenase
genes (i.e.,
hup
,
hym, hyc, ech and vhu
), as well as other genes indirectly associated with
reductive dechlorination. For example, the requirement for corrinoid cofactors to perform
reductive dechlorination suggests that monitoring the expression of genes encoding proteins
for corrinoid transport or salvage may serve as a proxy for monitoring actively dechlorinating
Dhc
populations.
Table
2.7
shows qPCR primers or probes for commonly utilized gene targets for the
analysis of
Dhc
and
Dhc
relatives. The quantitative analysis of biomarker genes (i.e., DNA)
provides useful information about the presence and temporal dynamics of the population of
interest. Although temporal analysis provides some clues about
Dhc
growth and activity, the
DNA-based analysis does not directly inform about activity (i.e., rates) and cannot distinguish
live and active cells from dead
Dhc
cells or free DNA released from lysed cells.
Targets that typically correlate more directly with activity are biomarker gene transcripts
(i.e., messenger RNA, or mRNA). A few studies have shown that the quantitative assessment
of biomarker mRNA provides information about activity under laboratory conditions (John-
son et al.,
2005
; Rahm and Richardson,
2008
). Although promising, this approach has
several drawbacks that limit its applicability, in particular when working with field samples.
RNA is inherently unstable and prone to degradation. The use of internal standards to
quantify RNA loss and RNA stabilizing agents can improve the analysis but uncertainties
remain especially when applying these techniques to natural populations (Johnson et al.,
2008
;
Ritalahti et al.,
2010b
).





















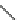




























Search WWH ::

Custom Search