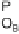Information Technology Reference
In-Depth Information
Figure 4.5
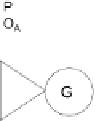
Wire OR-ing the outputs of two inverters yields a NAND gate.
given promoter. Second, two inverters form a NAND gate by “wiring-OR”
their outputs (Figure 4.5). These two features combine to provide a modular
approach to logic circuit design of any finite complexity, as described below.
Modularity in Circuit Construction
The modularity in biocircuit design stems from the ability to designate almost
any gene as the output of any logic gate. Consider a logic element
A
consisting
of an input mRNA,
M
A
, that is translated into an input protein repressor
R
A
,
acting on an operator,
O
A
, associated with a promoter,
P
A
. Let
P
A
be fused to
a structural gene,
G
Z
, coding for the output mRNA,
M
Z
. Figure 4.3 illustrates
these genetic elements. The DNA basepair sequence
G
Z
(or the corresponding
output mRNA sequence
M
Z
) that codes for an output protein,
R
Z
, determines
the gate connectivity because the output protein may bind to other operators
in the system. The specific binding of
R
Z
to another downstream operator,
O
Z
, connects gates because the level of
R
Z
affects the operation of the down-
stream gate.
To a first approximation, the choice of the sequence
G
Z
does not affect the
transfer function,
I
, of the inverter,
M
Z
=
T
◦
C
◦
L
(M
A
)
. An exception to
this rule occurs when
G
Z
codes for a protein that interacts with operator
O
A
or with input protein repressor
R
A
. Thus, the designer of
in vivo
logic circuits
must ensure that the signal proteins do not interact with circuit elements other
than their corresponding operators. The circuit designer should experimentally
verify this required protein noninterference before circuit design.
1
Any set of
non-interacting proteins can then serve as a library of potential signals for
constructing an integrated circuit.
Once protein noninterference is established, modularity of the network de-
sign affords a free choice of signals. Any suitable repressor protein and its
corresponding mRNA is a potential candidate for any signal, where the issue
of suitability is discussed later in this chapter. This modularity is necessary for
implementing a
biocompiler
—a program that consults a library of repressor
1
For example, the following simple
in vivo
experiment checks whether a protein affects a particular promoter.
First, fuse a fluorescent protein to a promoter of interest and quantify the
in vivo
fluorescence intensity. Next,
add a genetic construct that overexpresses the protein of interest. Finally, check the new fluorescence intensity
to determine whether the protein affects transcription.