Information Technology Reference
In-Depth Information
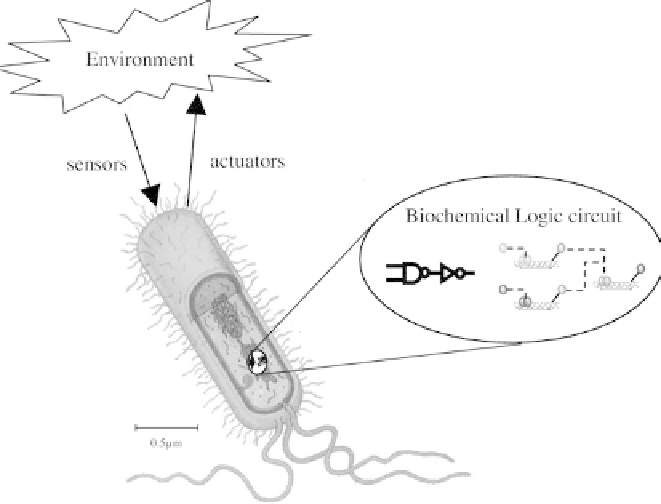
Figure 4.1
Embedding biochemical logic circuits in cells for internal computation
and programmed intercellular communications, extending and modifying the behavior
of cells and cell aggregates.
pharmaceutical synthesis, programmed therapeutics, and as a sophisticated tool
for
in vivo
studies of genetic regulatory networks. These applications require
synthesis of sophisticated and reliable cell behaviors that instruct cells to make
logic decisions based on factors such as existing environmental conditions and
current cell state. For example, a cell may be programmed to secrete particular
timed sequences of biochemicals depending on the type of messages sent by
its neighbors. The approach proposed here for engineering the requisite pre-
cision control is to embed internal computation and programmed intercellular
communications into the cells (Figure 4.1). The challenge is to provide ro-
bust computation and communications using a substrate where reliability and
reproducible results are difficult to achieve.
Biological organisms as an engineering substrate are currently difficult to
modify and control because of the poor understanding of their complexity.
Genetic modifications to cells often result in unpredictable and unreliable be-
havior. A single
Escherichia coli
bacterial cell contains approximately 10
10
active molecules, about 10
7
of which are protein molecules. The cumulative
interaction of these molecules with each other and the environment determines
the behavior of the single cell. Although complex, these interactions are not
arbitrary. Rather, cells are highly optimized information-processing units that