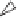Information Technology Reference
In-Depth Information
unscrambling in ciliates [6-9]. We study these operations from a computational
point of view. We namely summarize results in Kari and Kari [6] showing that, if
the recognition of identical pointers is assumed to be sufficient to trigger recom-
bination, the computational potential achieved is only that of finite automata.
We define circular words over
Σ
by declaring two words to be equivalent
iff one is a cyclic permutation of the other. In other words,
w
is equivalent to
w
iff they can be decomposed as
w
=
uv
and
w
=
vu
, respectively. Such a
circular word
•
w
refers to any of the circular permutations of the letters in
w
.
Σ
•
Denote by
the set of all circular words over
Σ
.
DEFINITION 1 If
x
∈ Σ
+
is a pointer, then the recombinations guided by
x
are
defined as follows:
u
xv
⇒
uxv
+
u
xv
(linear/linear)
uxv
+
(1)
uxvxw
⇒
uxw
+•
vx
(linear/circular)
(2)
u
xv
⇒ •
uxv
u
xv
(circular/circular).
•
+•
uxv
(3)
(See figures in Kari and Kari [6].) Note that all recombinations in definition 1
are reversible; the operations can be performed also in the opposite directions.
For example, operation (2) models the process of intramolecular recombi-
nation (Figure 10.2). After the pointer
x
finds its second occurrence in
uxvxw
,
the molecule undergoes a strand exchange in
x
that leads to the formation of
two new molecules:
uxw
and a circular DNA molecule
•
vx
. Intramolecular
recombination accomplishes the deletion of either sequence
vx
or
xv
from the
original molecule
uxwxv
and the positioning of
w
immediately next to
ux
.
u
x
v
u
v
+
x
x
x
v'
u'
u'
v'
+
u
x
v
x
v'
u'
Figure 10.2
Linear/linear recombination.