Biology Reference
In-Depth Information
Start
t
1
t
2
t
4
Intra-
cellular
t
3
Secreted
t
6
t
5
e
3
e
4
e
6
e
1
e
5
e
2
D2
D3
D1
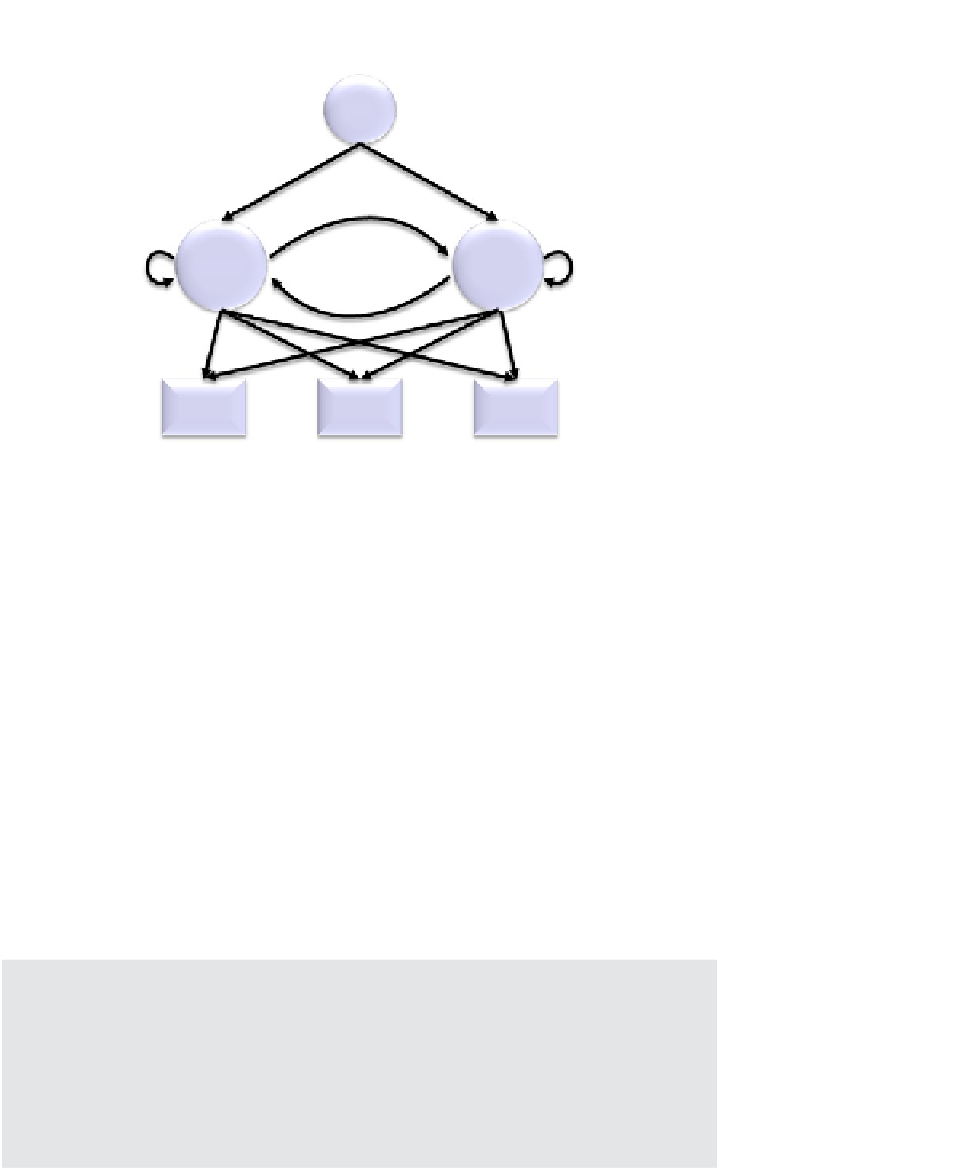
FIGURE 2.7
A very simple HMM to determine whether a protein is secreted or intracellular, based upon its
sequence of domains, D1-D3. Variables
t
1
-
t
6
are transmission probabilities;
e
1
-
e
6
are
emission probabilities.
There are several variants on the basic HMM, with slightly different
functionality.
HMMs have recently been used for tasks such as detecting genomic islands,
frequently associated with virulence, in newly sequenced bacterial genomes
(
Langille
et al.
, 2010
); identifying and classifying secretome proteins (
Craddock
et al.
, 2008
), and promoter prediction in the
Chlamydia
genome (
Malios
et al.
,
2009
). HMMs have long been used for gene finding (
Lukashin and Borodovsky,
1998; Azad and Borodovsky, 2004; Besemer and Borodovsky, 2005
), including
specifically identifying gene starts in microbial genomes (
Besemer
et al.
, 2001
).
A powerful approach to some problems is to combine HMMs with other bioin-
formatics tools. For example, (
Snir and Tuller, 2009
) combined an HMM with a
Maximum Likelihood approach in order to analyse and model phylogenetic
networks.
Software Availability
HMMER (
http://hmmer.org/
);
PSI-BLAST (
http://www.ncbi.nlm.nih.gov/Education/BLASTinfo/psi1.htm
).
PFTOOLS (
http://www.isrec.isb-sib.ch/profile/profile.html
)
.
GeneMark:
http://opal.biology.gatech.edu/GeneMark/
.