Biology Reference
In-Depth Information
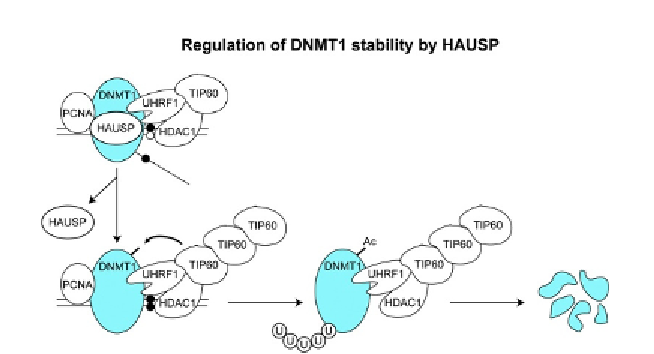
Figure 1.6 Regulation of DNMT1 stability by HAUSP. DNMT1 and associated proteins at
the replication fork function to maintain CpG methylation (transition from open circle to
filled circle) following DNA replication. Subsequent dissociation of HAUSP from the com-
plex of DNMT1 and interacting proteins, including UHRF1 and TIP60, leads to modifica-
tions of DNMT1 and eventually to its degradation. Details are discussed in the text.
overexpression of Tip60, the acetylation levels of H2AK5 are reduced. Fur-
thermore, downregulation of UHRF1 and DNMT1 resulted in a dramatic
decrease in the levels of H2AK5 acetylation, suggesting that UHRF1 and
DNMT1 together regulate H2AK5 acetylation via modulation of Tip60
(
Achour et al., 2009
).
Interactions between DMAP1 and DNMT1 regulate DNMT1's activ-
ities (
Mohan et al., 2011
). The known DMAP1 interaction domain is the
amino terminus of DNMT1 (118 amino acids in mouse and 125 amino acids
in human), although other region(s) of DNMT1 also interact with DMAP1.
Using
in vivo
genetic experiments to regulate the levels of DMAP1 protein
and forms of DNMT1 (one with the known DNMT1 interaction domain
and one without it), we showed that a sudden increase in “free” DMAP1 or
DMAP1 not interacting with the known DMAP1 interaction domain of
DNMT1 in preimplantation embryos resulted in the loss of a significant por-
tion of inherited imprinted methylation. Because DNMT1 is absolutely
required for maintenance of genomic imprints, these observations suggest
that excess or “free” DMAP1 interferes with the normal regulation of
DNMT1 by DMAP1. How DMAP1 regulates DNMT1 is not clear,
although we can propose two plausible models: a direct interaction between
DMAP1 and DNMT1 proteins or an indirect interaction via the interaction
between the Tip60 complex (of which DMAP1 is a member) and DNMT1.
Search WWH ::

Custom Search