Biomedical Engineering Reference
In-Depth Information
of 5 [67,68] and yield drug candidates more suitable for later medicinal chemistry
optimizations (Figure 11.11).
11.2.2.2 DNA-Routed Libraries
In 2004, Halpin and Harbury established another
milestone in DNA-encoded chemical library research [59,69]. For the first time
they developed a variation of the split-pool library construction in which the DNA
appendage served not only as a PCR-amplifiable identification tag but also as a code
for programming the stepwise library assembly.
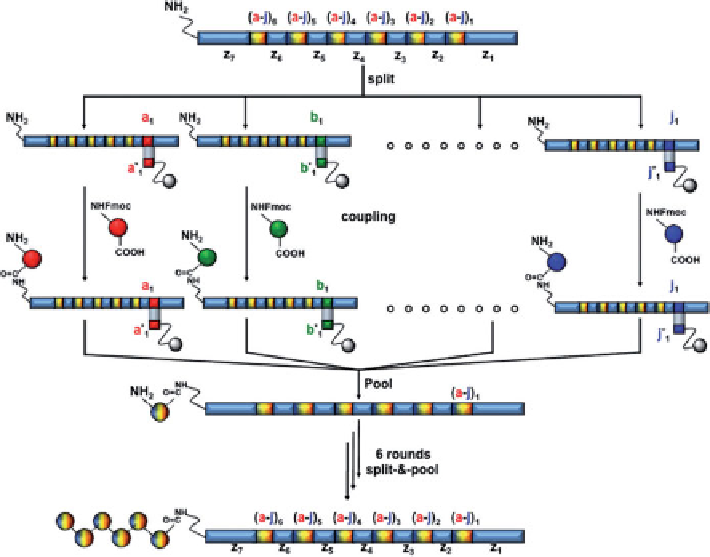
As illustrated in Figure 11.12, different oligonucleotide templates are physi-
cally separated (“routed”) by flowing into sequential cartridges containing appro-
priate anticodon DNA sequences, and processed in discrete chemical reactions [69].
Subsequently, the subpopulations are combined and flown through a second set of
FIGURE 11.12
DNA-encoded chemical library by DNA routing. An initial combinatorial
library comprising 10
6
different 340-mer-oligonucleotide templates was generated by PCR
assembly of smaller oligonucleotides. Each oligonucleotide contains six coding regions, each
encoding for 10 different amino acids [(a to j)
1-6
], and seven constant domains (z
1-7
). The tem-
plate library was split using complementary oligonucleotides bound on affinity chromatography
resins [(a* to j*)
1-6
] (DNA routing). Following conjugation to the corresponding Fmoc-amino
acid and deprotection, oligonucleotides were eluted and the initial template library recon-
stituted. Six rounds of DNA routing yielded to the final library of 10
6
different
N
-acylated
pentapeptides. (
See insert for color representation of the figure.
)