Biomedical Engineering Reference
In-Depth Information
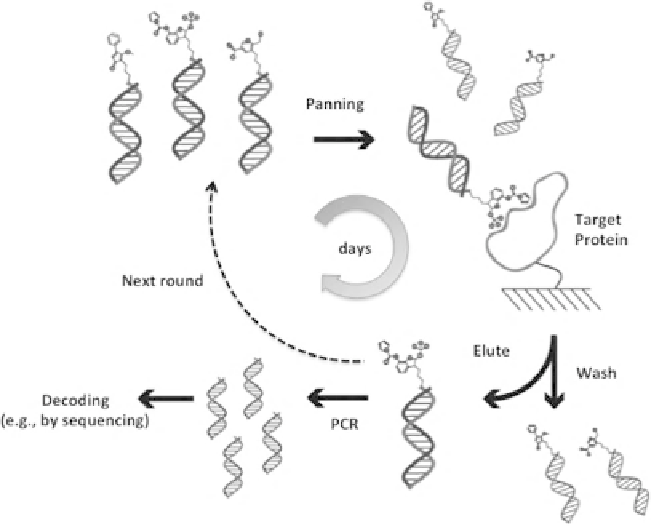
FIGURE 11.3
DNA-encoded chemical library panning. Initially, the DNA-encoded chem-
ical library is incubated with a target protein of choice immobilized on a solid support.
Following the removal of nonbinders by washing cycles of the solid support, DNA tags of the
retained library members are PCR-amplified. Eventually, decoding (e.g., by DNA sequencing)
and deconvoluting the enriched DNA codes reveals the identity of the compounds selected.
In principle, the panning cycle can be iterated multiple times (dashed arrow), yielding fur-
ther enrichment of binding library members over nonbinding compounds. The process can be
performed in just a few days, and multiple targets can be selected in parallel.
Drawing an analogy between conceptually similar display technologies, the
authors postulated that DNA-encoded small molecules could be selected in vitro
against virtually any target protein of choice [47]. As already popularized by other
polypeptide-based technologies and in resemblance to the gold-mining process of
washing gravel in a pan to separate gold from contaminants, the selection procedure
was termed
panning
.
In short, after imposing a selection pressure (e.g., incubating the DNA-encoded
library with the target protein of interest immobilized on a solid support), washing
cycles allow for the enrichment of specific binding moieties whose DNA codes are
eventually PCR-amplified and decoded (e.g., by DNA sequencing, Figure 11.3).
Relative quantification of library members before and after the selection process
leads to the identification of enriched small-molecule structures. In principle, such a
panning procedure can be repeated multiple times, yielding to further enrichment of
binding library members over nonbinding compounds (Figure 11.3, dashed arrow).