Biomedical Engineering Reference
In-Depth Information
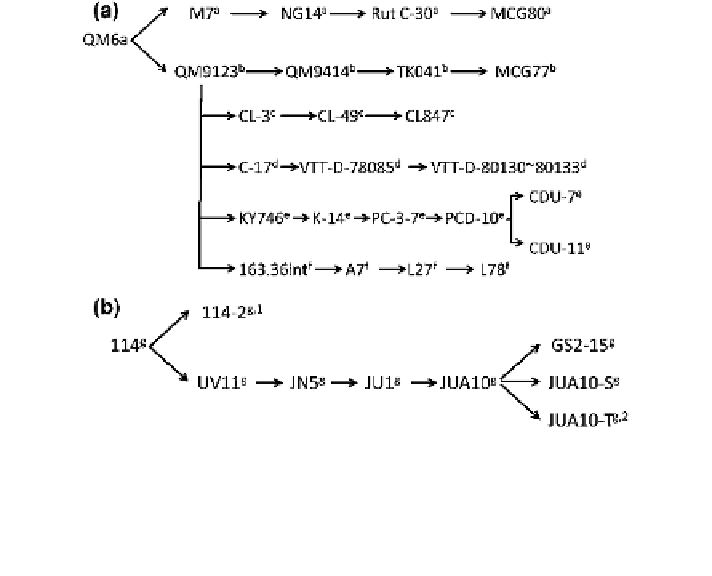
Fig. 5 The genealogy of Trichoderma reesei (a) and Penicillium decumbens (b). a-g strains
isolated/constructed by Rutgers University, USA; the US Army Natick Laboratories; Cavla
Laboratories, France; VTT Technical Research Centre of Finland; Kyowa Hakko Kirin, Japan,
formerly Kyowa Hakko Kogyo, Japan; Chiron Corporation, formerly Cetus Corporation, USA;
the State Key Laboratory of Microbial Technology, Shandong University, China. 1 Filter
paperase unit (FPU) 0.35 IU/mL, 2 FPU 18.9 IU/mL
The genealogy of the representative strains created in our laboratory with the
aforementioned approaches is presented in Fig.
5
b. Large numbers of mutant
strains that exhibit a diversity of characteristics were obtained through UV and
N-methyl-N
0
-nitro-N-nitrosoguanidine treatment, and various other techniques.
P. decumbens strains JU15 and JU1, which are resistant to catabolite repression,
were isolated using a culture medium containing glucose [
67
].
Aside from these traditional mutation techniques, genetic engineering was also
exploited to improve the expression levels of cellulases and hemicellulases in
fungi. For example, Liu et al. [
102
] modified the promoter region of cbh1 and
alleviated the glucose repression effects by deleting the binding sites of the CreI
protein, as well as repeatedly inserting the CCAAT box and the ACE II binding
site into the modified cbh1 promoter in T. reesei. Su et al. [
98
] obtained higher
cellulase activity in T. reesei by reconstructing the effector domain of ACE II.
Zhang et al. [
103
] replaced the promoter of the gene encoding bG with a four-copy
cbh1 promoter in T. reesei. The recombinant strain showed a significant increase
in bG activity, filter paper activity, and better saccharification of corncob residues.
A similar approach was utilized by Wang et al. [
104
], who created a T. reesei
strain with higher CMCase activity by replacing the coding sequence of CBH I
with that of EG III. The genome sequencing of P. decumbens was recently
completed (unpublished data), and will provide insight into the role of cellulase
genes and the regulatory mechanisms of cellulose degradation. To improve gene
Search WWH ::

Custom Search