Biomedical Engineering Reference
In-Depth Information
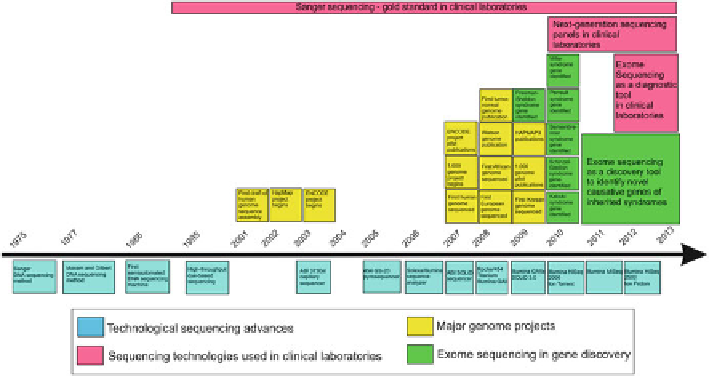
Fig. 1.1
Timeline of the major advances in sequencing technologies starting with Sanger
sequencing and ending with the next-generation-sequencing (
blue
). The major genome projects
that were made possible by these methods are shown (
yellow
). In addition to clinical exome
sequencing, the timeline of Sanger sequencing as a gold standard in clinical laboratories is depicted
(
purple
). The uses of NGS in exome sequencing project with gene discovery objectives have also
been noted (
green
)
followed by specifi c genetic disorder examples where NGS technologies have made
great advancement in the diagnosis of patients including prenatal diagnosis
(Chap.
5
)
, muscular dystrophies (Chap.
6
), hearing loss (Chap.
7
)
, and exome analy-
sis as applied to gene discovery and molecular diagnostics (Chap.
8
)
. There are
advantages and disadvantages to all technologies. In Chap.
9
, we will address the
challenges of NGS in molecular diagnostics.
1.2
Principle of Sanger Sequencing
In 1975, Sanger introduced his “plus and minus” method for DNA sequencing
(Fig.
1.1
; Sanger and Coulson
1975
). This was a critical transition technique lead-
ing to the modern generation of methods that have completely dominated sequenc-
ing over the past 30 years. The key to this advance was the use of polyacrylamide
gels to separate the products of primed synthesis by DNA polymerase in order of
increasing chain length (Hutchison
2007
). In Sanger dideoxy DNA sequencing, a
DNA-dependent polymerase is used to generate a complimentary copy of a single-
stranded DNA template, also known as sequencing by synthesis (SBS) (Sanger
1988
; Sanger et al.
1977a
,
b
). By beginning at the 3
end, a new chain of a primer
DNA complementary to a single-stranded “template” DNA strand is synthesized.
′
Search WWH ::

Custom Search