Biomedical Engineering Reference
In-Depth Information
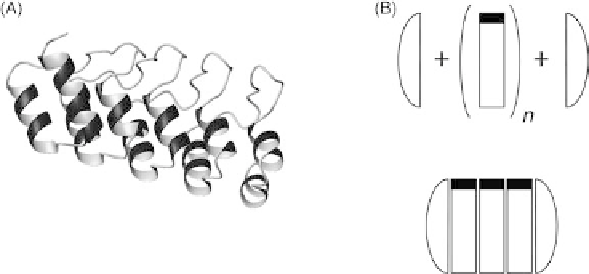
FIGURE 34.1
DARPins (designed ankyrin repeat proteins) [11]. (A) Structure of a DARPin shown
in ribbon representation (PDB-ID: 1MJ0). The molecule comprises several repeats that stack to form
one elongated domain. One repeat consists of a beta-turn, followed by a helix-loop-helix motif and a
loop connecting to the next following beta turn. Special hydrophilic terminal repeats (capping
repeats) shield the continuous hydrophobic core formed by the stacked central repeat modules. The
molecule has the shape of a hand, where the palm of the hand is typically involved in the target
interaction, and forms a large potential-interaction surface. The N term is at the left, the C term at the
right side of the picture. (B) Schematic concept of generating DARPin libraries starting from capping
repeats and varying numbers of a consensus-designed repeat modules. Randomized areas are
indicated by black bars. Below, a schematic drawing of an N3C molecule is shown (three repeat
modules flanked by an N- and a C-terminal capping repeat), exposing a large potential target-
interaction surface. An example of such an N3C DARPin is shown in (A).
corresponding to about a tenth of an IgG immunoglobulin
[4]. The flexibility of the repeat number is similarly seen in
other repeat protein proteins [6]. Nonrepeat protein target-
binding proteins typically have a fixed molecular size in the
range of 6-35 kDa depending on the fold [8].
The stability of natural binding proteins is often trimmed
to the biological requirements and can be diverse. For
example, some ankyrin repeat proteins are extremely stable,
comparable to designed ankyrin repeat proteins [18], while
others, which require higher turnover due to their biological
function, show low stability and fast turnover [19]. For most
natural binding proteins analyzed, simple protein engineer-
ing allows increasing the thermodynamic stability, indicat-
ing that the different folds are intrinsically stable [8].
Overall, nature has evolved a large number of binding
proteins that can serve as a source for alternatives to
antibodies.
one or several regions of the scaffold by library methods
(typically based on knowledge of the sites of interaction of
the scaffold), and (iii) selection of binding variants by state-
of-the-art selection technologies such as ribosome display or
phage display, and the characterization of the selected
binders.
Already the first step is crucial. To be able to efficiently
work with a scaffold, the protein should be expressable in a
simple host at high levels, allowing for simple production
and purification of variants. The scaffold should have a
sufficient buffer of stability allowing randomization of the
scaffold. In other words, the parental, nonrandomized scaf-
fold should certainly exhibit a high degree of stability. Also,
the protein should have a low potential to be immunogenic.
The next step classically consists of analyzing the natural
binding site of the scaffold using literature or own exper-
imental data, designing a randomization scheme, and gen-
erating a library of variants by DNA randomization methods.
The better the understanding of the natural target-interaction
of the scaffold is, the easier the design gets. The availability
of 3D structures of the scaffold in complex with a target
molecule is a great plus. In any case, the randomization
scheme also has to be adapted to the subsequent selec-
tion/design process.
Typically, the DNA library of a scaffold is then trans-
formed to the ribosome-display [22], phage-display [23], or
yeast-display [24] format, and binding proteins are gener-
ated using these methods. Depending on the scaffold, the
design, and the selection methods, high affinities, and high
34.2.3 Tailoring Nonantibody Binding Proteins
Over the past 20 years, many of the naturally occurring
binding proteins have been tested to serve as alternatives to
antibodies. The list comprises proteins such as repeat pro-
teins (see below) [20], fibronectin [7], protein Z [21], lip-
ocalins [14], SH2, SH3, PDZ,
g
-crystallin and others
(reviewed, for example, in Binz et al. [8]). Typically, gener-
ating alternatives to antibodies involves (i) the choice of a
scaffold (in this context scaffold refers to the protein which
is used as a matrix for the following), (ii) randomization of
Search WWH ::

Custom Search