Biomedical Engineering Reference
In-Depth Information
Phage expressing mTCR on its surface
Phagemid carrying genes
for mTCR
α
-
and
β
-
chains
Individual CDR libraries panned against peptide-MHC
mTCR with a million fold-increased affinity
t
¾
= 1135 min
t
¾
= 15 min
t
¾
= 6.4 sec (wt)
Time
Affinity-enhanced mutants selected
Mutated CDRs combined
Tested on Biacore
Proteins made by
E. coli
expression and refolding
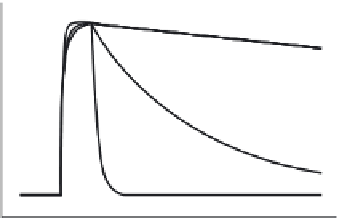
FIGURE 32.3
Affinity maturation of mTCRs with phage display. Mutations are being introduced
into the CDR regions of the
a
and
b
chains. Expressed on phage, the individual CDR mutants are
panned against the specific peptide-MHC. This typically increases the affinity from micromolar to
nanomolar range. At the next stage, the optimum CDR sequences are brought together in mTCR
format, the chains expressed in E. coli, refolded in desired combinations and the affinity is tested on
the Biacore. This is an iterative process and enables the enhancement of the affinity into the
picomolar range.
chain is encoded as a free (nonfused) chain. The resulting
phage particles display the fused TCR
b
chains paired with
the secreted
a
chains, disulfide-linked to each other. PCR
mutagenesis is used to introduce variability to the targeted
regions (one CDR at a time), and the libraries are panned
on the relevant immobilized peptide-MHC. Creation of
second-generation phage TCR libraries, and repeated
selections against decreasing concentrations of antigen,
result in isolation of TCRs with further incremental
improvements in affinity. At this stage, a number of
selected mutants are made as soluble mTCRs through
E. coli expression and in vitro refolding, and their affinities
measured using a surface plasmon resonance microchip-
based detector (Biacore). mTCRs featuring mutations in
just one or two of their CDRs generally exhibit affinities in
the nanomolar range, the average increase being
mutations in several CDRs are combined, usually generat-
ing mTCR variants exhibiting affinities in the picomolar
range. Thus, the phage display selection process typically
produces affinity enhancements of a million fold com-
paredtotheoriginalnative TCR. Examples of mTCRs
affinity matured by this approach are shown in Table 32.2.
The high-affinity TCRs can be labeled, for example, with
a fluorescent molecule, and used to visualize or quantitate
the specific antigen on cells (Figure 32.5A). To our
knowledge, these high-affinity mTCRs constitute the first
tool that has enabled direct detection of cell-surface
peptide-MHC, an application that has been used in vac-
cine validation and which could potentially be used for
future diagnostic purposes [26,27]. No detectable levels of
nonspecific binding to irrelevant peptides or antigen neg-
ative cells have been observed with any of the soluble
mTCRs listed in Table 32.2.
1000-
fold. Ultimately,
selections of
the most promising
















Search WWH ::

Custom Search