Biology Reference
In-Depth Information
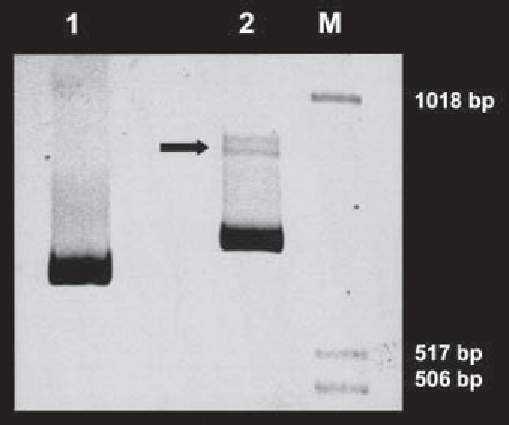
Fig. 1. Six percent PAGE-analysis of PCR amplified COMP cDNA. Total RNA
was isolated from a skin fibroblast cell line established from a patient with pseudo-
achondroplasia and reverse transcribed into cDNA with both oligo-dt and pd(N)
6
primers as described.
Lane 1,
COMP C-terminal domain cDNA PCR-amplified by
primers HCt-F and HCt-R (702 bp).
Lane 2,
COMP Type III repeat domain cDNA
PCR-amplified by primers HT3-F and HT3-R (789 bp).
M,
molecular weight marker.
A white arrow indicates a putative heteroduplex. Sequence analysis of clones
confirmed that this patient was heterozygous for a COMP gene mutation.
a. 94
°
C for 1 min (template denaturation).
b. 55
°
C for 1 min (primer annealing).
c. 72
°
C for 1 min (template elongation).
2.
After cycle is complete, remove 15
L of DNA loading
buffer and analyze on a 6% polyacrylamide gel in 1X TBE buffer (
see
Note 3
).
Stain the gel with ethidium bromide (10
µ
L of PCR product, add 5
µ
µ
g/mL) and view DNA with an UV
transilluminator (
Fig. 1
and
Fig. 2
).
3.2.2. SSCP Analysis of Genomic DNA Amplified by PCR
1.
Remove 2
µ
L of PCR product to a fresh tube, add 10
µ
L of denaturing DNA
loading buffer and heat samples to 95
°
C for 2 min. Place denatured DNA on ice
for 2 min.
2.
Load sample onto a 6% polyacrylamide gel precooled to 4
°
C and run at 300 V for
3-4 h (
see
Note 4
).
3.
After the run is complete, separate the plates gently, remove the gel, and place in
500 mL of gel-fixative solution for 20 min.
4.
Wash the gel twice in dH
2
O for 5 min.

Search WWH ::

Custom Search