Biology Reference
In-Depth Information
A
B
3
10
G
A
C
U
G
U
U
C
A
C
C
U
C
U
G
A
C
A
A
G
U
U
UU
G
C
A
A
A
C
U
3
10
G
U
G
G
A
U
A
C
C
U
C
A
C
C
U
A
C
G
C
U
C
A
G
G
C
G
A
G
U
U
C
G
U
U
U
G
G
30
U
P2
P2
P1
P1
100
100
A
U
G
G
30
U
G
G
5
5
HO
P3
HO
P3
G
G
C
C
U
CG
C
G
G
C
C
C
CG
U
20
A
G
U
C
A
A
A
C
P1.1
P1.1
J4/2
20
J4/2
G
C
A
A
U
G
C
U
G
U
G
C
U
C
C
U
A
C
G
A
C
A
G
G
G
A
C
A
C
C
C
A
C
C
A
C
U
C
G
C
A
U
U
C
U
U
C
A
C
C
94
95
40
40
90
P4
90
P4
50
50
80
80
60
60
A
70
A
G
A
A
A
U
U
C
A
G
G
U
U
70
C
1
Consensus
Figure 4.10 Secondary structures of (A) the drz-Ppac-1 and (B) drz-Cjap-1 ribozymes. (C)
Alignment of 80 drz-Cjap-1 sequences. The coloring of the consensus alignment is
identical to that seen in the secondary structure, with 10 nt upstream and downstream
of the ribozyme also shown (black). Gaps in the consensus sequence correspond
to positions at which a single copy of the ribozyme contains a nucleotide insertion.
ribozyme. Such observations are consistent with a model in which the drz-
Cjap-1 sequences perform a role in retrotransposition comparable to other
non-LTR-associated ribozymes, likely acting as a 5
0
cap for the RNA and
helping to initiate translation.
126




























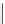



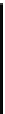












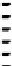


































Search WWH ::

Custom Search