Biology Reference
In-Depth Information
h1
s1
h2
h3
h4
h5
s2
h4´
s3
h3´
h1´
h5´
s4
h6
s5
h6´
s6
h2´
s1
3
h1 0:0 GNNNNNN:NNNNNNY
h2 0:1 NNNNNNN:NNNNNNN
h3 0:0 NNN:NNN
h4 0:0 U:G
h5 0:1 NY:RN
h6 0:0 NNNN[26]:[26]NNNN
N
N
N
N
N
N
N
N
N
N
N
N
N
N
N
N
N
N
N
N
h2
h1
N
N
N
N
G
Y
h3
NN
NN
NN
5
s2
s3
RY
NN
N
C
G
N
h5
A
R
N
C
U
s1 0 NN******
s2 0 N
C
s3 0 N
s4 0 N****
s5 0 NNN[30]
s6 0 CNRA*
H4
s4
s6
N
N
N
N
N
N
h6
s5
Figure 4.6 A typical RNABOB descriptor. Each motif that forms the secondary structure
is individually defined on the left, and their relation to one another is specified in the
single line appearing at the top of the figure. In the descriptor, an
“
h
”
designation cor-
responds to a helical region, and an
“
s
”
designation corresponds to a single-stranded
portion of the molecule. When defining a helix, a number to the right of the colon indi-
cates the number of allowed mismatches to that helix, whereas a number to the left or
following an
“
s
”
designation defines the number of allowed variations to a defined
sequence. Asterisks or bracketed numbers in the descriptor specify the maximum num-
ber of additional nucleotides a region can contain (e.g., s1 is at least two nucleotides but
can contain as many as eight nucleotides).
search for regions of complementarity between any portions of the genome
that are spaced appropriately to produce the gross structure of the motif.
119
Following a genomic search, RNABOB outputs a file that contains all
sequences capable of assuming the defined structure with each region
defined in the descriptor individually delimited. This allows for easy parsing
into spreadsheets, where the data can be further organized according to a
particular region, such as the highly conserved core of the HDV ribozyme.
Since optimal descriptors often allow for various mutations and insertions
that independently may not disrupt proper folding but collaboratively lead
to inaccurate structures, organizing the data in such a way allows suboptimal
sequences to be quickly eliminated from the pool of candidate motifs.
5.7. HDV-like ribozymes found using RNABOB
A genome-wide search for sequences capable of assuming an HDV-like
structure using the RNABOB software revealed a plethora of
in vitro
and
in vivo
active ribozymes in a wide range of organisms. The first HDV-like
ribozymes identified through structure-based searches included several


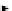




























Search WWH ::

Custom Search