Biology Reference
In-Depth Information
ssDNA
Ligation and
primer extension
Isolation of ssDNA
6
1
Linear
dsDNA
Circular
dsDNA
Reverse transcription
and PCR
Rolling circle
transcription
5
2
Dimeric RNA
Uncleaved, concatameric RNA
Isolation of dimers
In vitro
self-cleavage
4
3
Figure 4.4 Selection scheme for the in vitro isolation of self-cleaving ribozymes.
A single-stranded DNA pool containing a T7 promoter region (blue), constant primer
regions (red and orange), and a stretch of random nucleotides (black) is (1) ligated
and primer extended, followed by (2) rolling-circle transcription to yield concatemeric
RNA molecules. These RNAs are then (3) incubated in buffer containing Mg
2þ
, wherein
active sequences will self-cleave from concatemers to monomers (cleavage sites indi-
cated by triangles). Dimers, which will contain both the forward and reverse primers in
proper orientation, are then (4) isolated and (5) subjected to reverse transcription and
PCR. The DNA is then (6) denatured to yield the full-length single-stranded DNA for use
in subsequent rounds of selection.
5.3. Structural and enzymatic characteristics of the CPEB3
ribozyme
A genomic selection for self-cleaving ribozymes is capable of revealing all
self-cleaving sequences in a genome active under the selection conditions,
but it does not provide any information as to the type of motif that is respon-
sible for the reaction. To achieve the latter objective, extensive characteri-
zation of the identified sequences is required. Consequently, the secondary
structure is only known for the motif residing in the
CPEB3
gene, although
four sequences capable of self-scission were found in the human genome.
2
The
CPEB3
ribozyme possesses several of the hallmark features associ-
ated with HDV ribozymes.
CPEB3
is able to catalyze its self-scission in a
wide range of divalent metal
ions including Mg
2
þ
,Mn
2
þ
,Ca
2
þ
, and






































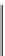


Search WWH ::

Custom Search