Biology Reference
In-Depth Information
A
B
U20
U20
O
O
N
O
P
O
N
H
2
N
HN
P
O
−
O
C22
H
2
N
HN
O
O
−
NH
O
C22
NH
O
N
O
p
K
a
~6.2-6.4
H
N
U75
N
O
O
G25
O
HOH
O
−
N
C75
G25
O
O
O
−
O
H
N
N
HN
P
O
N
M
O
O
N
N
N
P
M
HO
O
N
H
HOH
NH
O
O
HO
O
N
O
-
O
O
NH
HO
O
-
O
O
P
N
N
NH
2
O
N
O
P
O
O
N
N
NH
2
HN
O
O
O
G1
O
O
N
G1
U-1
O
P
OH
HN
O
O
O
P
OH
O
O
O
−
U-1
O
−
O
O
−
O
P
O
O
3
ribozyme
OH
O
5
leader sequence
O
3
ribozyme
5
leader sequence
C
U20
O
O
N
P
HN
H
2
N
O
-
O
C22
NH
O
p
K
a
~5-5.5
H
N
O
C75
N
O
G25
-
O
H
O
N
O
N
N
P
N
O
HO
N
O
O
NH
HO
N
NH
2
N
O
G1
O
P
OH
O
O
−
O
3
ribozyme
Figure 4.3 Active-site structures of the genomic HDV ribozyme. (A) Model of the C75U
variant in precursor form.
31
(B) Model of the wild-type C75 with inhibited substrate
strand lacking the U-1 2
0
hydroxyl nucleophile.
33
The pK
a
of C75 was measured using
Raman crystallography.72. (C) Active-site groups in the product form of the ribozyme
with the pK
a
of C75 measured by NMR.
33,73
The substrate strand is colored blue, the
active-site metal ion is green, and the catalytic nucleobase is red. The nucleophile, phos-
phorus of the scissile phosphate, and the 5
0
oxygen of the leaving group are also colored
red.
to that used for structure determination by X-ray crystallography, affording a
rare combination of spectroscopic and diffraction data on nearly the same
molecule.
36
When the protonation of the C75 nucleobase in the self-cleaved
ribozyme was measured using solution NMR, the p
K
a
was estimated to be
higher than that for free or base-paired cytosine, but not as high as
the apparent p
K
a
of the reaction.
76
The authors concluded that while the
active site of the product caused a shift in C75 p
K
a
, protonation of the
nucleobase at neutral pH must be associated with an earlier step in the mech-
anism. Attempts tomeasure the p
K
a
byNMR in several inhibited forms of the











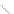


















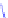



























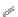



















































































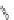

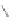










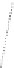
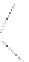





















































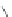


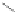










































Search WWH ::

Custom Search