Agriculture Reference
In-Depth Information
Biomphalaria
O. frivaldszkyi
O. compromissus
O. b. bihariensis
O. b. rendzinicola
O. exacystis
O. aporus
O. permagnus
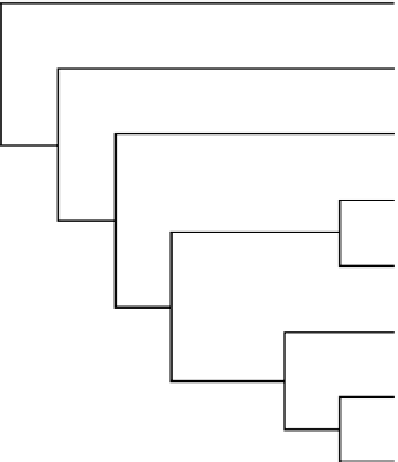
FIGURE 7.6
Maximum parsimony tree from six
Octodrilus
species based on COI analysis.
Discussion
Moreover, in the 16S sequences, our data ranged between 474 and 500 bp and seem to be more
complete than others reported. In the COI sequences, our data ranged between 650 and 672.
The very low homoplasy index of 0.0573 (required theoretic level under 0.5) proves that the
maximum parsimony tree presented here for ribosomal fragments of 16S rDNA is correct. Never-
theless, the 6.51% parsimony informative characters are quite low. Comparatively, the 22.01% of
parsimony informative characters and a homoplasy index of 0.2247 suggest a higher taxonomic
discriminatory value of the fragments of mitochondrial CO (COI).
Thus, at this stage of our research, the image of the cladogram for 16S (
Figure 7.5
) seems to
be less informative than that for COI (
Figure 7.6
). The clustering of species in both cladograms
shows the existence of the same number of taxa as differentiated by single- (
Figure 7.3
)
and
multiple-character
(
Figure 7.4
)
numerical analyses.
To answer the basic questions of how many
Octodrilus
species occur in the Carpathians; the
hypothetical and empirical trees from Figures 7.3 to Figure 7.6 are analyzed.
First, the three giant species were placed on different branches, which suggests that they are
really different taxa. The distance between
O. aporus
and
O.
permagnus
is relatively small, in
accordance with the original description, but they differ clearly from
O. frivaldszkyi.
Second, the
position and distances among
O. bihariensis, O. compromissus,
and
O. exacystis
prove that they
are also different species.
The relative positions of the six
Octodrilus
species in the tree diagrams are different but resemble
their positions in the hypothetical single-character, step-by-step tree, or in multiple-character anal-
yses. For example, the grouping of
O. frivaldszkyi
with
O. bihariensis
and
O. compromissus
and
of
O. exacystis
with
O. aporus
and
O. permagnus
fit the patterns obtained by numerical taxonomy.
The subspecies of
O. bihariensis
show very close relatedness.
In conclusion, molecular taxonomy research confirms or validates the species that were iden-
tified and described by numerical taxonomy. Species clustering and branching indicate possible
phylogenetic relationships.