Biology Reference
In-Depth Information
A
B
15,000
10,000
Mammals
Zebrafish
dnmt1
dnmt1
dnmt3
dnmt4
dnmt5
dnmt6
dnmt7
dnmt8
DNMT1
DNMT2
DNMT3A
DNMT3B
1200
1000
800
600
400
200
dnmt3
dnmt4
dnmt5
dnmt6
dnmt7
dnmt8
0
Oo
1-c
16-c
128/
256-c
MBT
Post-
MBT
C
D
Promoter methylation
Sperm
7000
6000
MBT
4000
2000
1-2 cell
3h
5h
Sperm
Pre-
MBT
MBT
Post-
MBT
Methylated in sperm, pre-MBT, MBT, and post-MBT embryos
Methylated in sperm only
Methylated in pre-MBT, MBT, and post-MBT embryos
Methylated in MBT and post-MBT embryos
Methylated in post-MBT embryos only
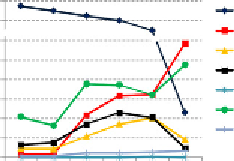
Figure 3.2 DNA methylation patterns in the zebrafish embryo. (A) dnmt gene
orthologs in mammals and zebrafish. (B) Expression patterns of zebrafish dnmt genes
during pre-MBT, MBT, and post-MBT development. Graph depicts mRNA detection
levels from our RNA-seq data (
Aanes et al., 2011
) in unfertilized eggs (Oo), and 1-cell
(fertilized), 16-cell, 128-/256-cell, MBT, and post-MBT (4.5-h postfertilization) embryos.
(C) Global DNA methylation changes during early development. (D) Changes
in promoter DNA methylation levels in sperm and at pre-MBT (256-cell stage), MBT
(1- to 2000-cell stage), and post-MBT (4.5-h postfertilization) stages. Distinct shadings
indicate developmental stage-specific methylation.
3. DNA METHYLATION CHANGES DURING THE MZT
An outstanding question has long been how transcriptional activation
is regulated at the time of ZGA. Recent evidence from zebrafish and other
species indicates that establishment of the embryo's own gene expression
program involves a chromatin remodeling implicating DNA methylation
and demethylation, and histone modification mechanisms. How these