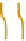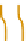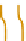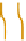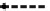Biology Reference
In-Depth Information
HDAC
HDAC
HP1
HP1
Me
Me
Me
G9a
DNMT3
G9a
H3K4
H3K9
H3K9
Ac
Ac
Ac
ON
OFF
OFF
Figure 2.5 Epigenetic regulation of the Oct-4 promoter during development. Transcrip-
tionally active Oct-4 promoter in pluripotent cells harbors acetylated histones, H3K4me,
and no CpG methylation (white circles). During differentiation of ES cells, the gene
promoter is silenced and undergoes deacetylation by histone deacetylases (HDAC),
methylation of H3K9 by methyltransferase G9a, and recruitment of HP1. Silencing is still
reversible until CpG methylation (red circles) catalyzed by DNMT3 locks the promoter in
an inactive state.
methylation at pluripotency genes, which, if incomplete, leads to partial
reprogramming (
Mikkelsen et al., 2008
).
How does DNA methylation maintain promoter silencing? One mech-
anism is that cytosine methylation inhibits the binding of transcription fac-
tors. Many transcription factors recognize sequence motifs that contain
CpGs (
Deaton & Bird, 2011
) and some of them show reduced binding affin-
ity when the recognition motif is methylated (
Campanero, Armstrong, &
Flemington, 2000; Iguchi-Ariga & Schaffner, 1989; Kim, Kollhoff,
Bergmann, & Stubbs, 2003
). Another model is that DNA methylation mod-
ifies the local chromatin by recruiting 5mC “readers.” Proteins that specif-
ically bind methylated DNA have been identified, such as those having a
methyl-CpG binding domain (MBD1, MBD2, MBD4, MeCP2) and those
recognizing 5mC with zinc-fingers (ZBTB4, ZBTB33, and ZBTB38)
(
Joulie, Miotto, & Defossez, 2010
). Most of these proteins interact with fac-
tors that deposit heterochromatin marks, which led to propose that they
mediate DNA methylation-dependent gene repression by inducing hetero-
chromatinization at sites of dense DNA methylation. Whereas there is evi-
dence for this model in cancer cells (
Lopez-Serra & Esteller, 2012
), their role
in translating the DNA methylation signal in normal cells still remains to be
further clarified because surprisingly little MBD target genes have been iden-
tified so far (
Joulie et al., 2010
). In addition, the several single and combined
knockout mice generated so far show no overt phenotype (
Martin
Caballero, Hansen, Leaford, Pollard, & Hendrich, 2009
), which is in clear
contrast with the lethality of DNMT-knockout mice.
There are numerous differences in DNA methylation between somatic
cell lineages, indicating that further changes in DNA methylation occur in