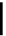Biology Reference
In-Depth Information
Zygote
2 Cell
4 Cell
8 Cell
16 Cell
Blastocyst
♀
♂
H3K9me2
♂
♀
H3K9me3
♂
♀
H4K20me2
♂
♀
H4K20me3
♂ ♀
Figure 1.2 Dynamics of the main constitutive heterochromatic marks during preim-
plantation development. All heterochromatin marks analysed to date are asymmetri-
cally localized on the maternal chromatin during the zygote stage. The levels of
H3K9me2 and H4K20me2 are maintained relatively stable during the early stages of
development, whereas global H3K9me3 levels decrease between the 4-cell and the
16-cell stage. H4K20me3 is undetectable on embryonic chromatin by the end of the
2-cell stage and similar kinetics have been described for H3K64me3, which is only pre-
sent during the zygote stage and is absent afterward.
H3K64me3
than what was originally thought and that DNA methylation in the zygote
occurs both actively (e.g., before the first round of replication) and passively
(concomitant with replication). DNA demethylation has been suggested to
be, at least partially, the consequence of the conversion of cytosine methyl-
ation to hydroxymethylation by the Tet proteins (
Gu et al., 2011; Inoue &
Zhang, 2011; Tahiliani et al., 2009; Wossidlo et al., 2011
). Together, this
results in an asymmetry in DNAmodifications between the paternal genome
(enriched in DNA hydroxymethylation) and the maternal genome
(enriched in DNA methylation) at the zygote stage (
Gu et al., 2011;
Inoue & Zhang, 2011; Iqbal, Jin, Pfeifer, & Szabo, 2011; Wossidlo et al.,
2011
). As development progresses, the differences in DNA methylation
and hydroxymethylation disappear (
Inoue & Zhang, 2011; Iqbal et al.,
2011
). The overall DNA methylation levels in the embryo decrease during
early development and then increase in the epiblast at the blastocyst stage
(
Mayer et al., 2000; Santos, Hendrich, Reik, & Dean, 2002
). On the
imprinted genes, however, DNA methylation is not completely lost and