Biology Reference
In-Depth Information
B
A
Sperm
0
Hypomethylated
promoters
Pre-MBT
0
MBT
0
Post-MBT
Methylated
promoters
0
Fibroblasts
0
H3K4me3
H3K27me3
10 kb
Chr.16
H3K9me3
Methyl-C
Non-methyl-C
Transcripts
skap2
hoxa9b
hoxa10b
hoxa11b
hibadhb
hoxa2b
hoxa13b
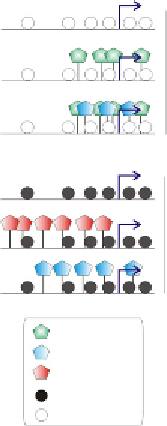
Figure 3.3 Differential marking of hypomethylated and methylated DNA regions by
modified histones in zebrafish embryos. (A) DNA hypomethylation of a developmentally
regulated gene cluster. Browser view of DNAmethylation profiles over a 54-kb fragment
of the zebrafish hoxa locus on chromosome 16. Note the hypomethylated state of the
locus in sperm and embryos, and local recurrent methylation in fibroblasts. (B) Differ-
ential association of H3K4me3 and H3K9me3 or H3K27me3 with hypomethylated
and methylated promoters, respectively, in pre-MBT embryos.
Nevertheless, evidence suggests that the hypomethylated state may create a
transcriptionally permissive “platform” for developmental gene expression.
Recent work from our laboratory based on differentiation of adipose
tissue-derived progenitor cells into adipocytes has shown that whereas meth-
ylation of lineage-specific promoters may be predictive of lineage restriction
(manifested by inability to differentiate into a specific lineage), the
unmethylated state, in contrast, has no predictive value on gene activation
and lineage differentiation (
Sørensen et al., 2010, 2009
). The absence of meth-
ylation may nonetheless enable a transcriptionally permissive state for later
gene activation, but potential for activation is likely modulated by other
mechanisms (such as histone modifications). Similarly, in early zebrafish
embryos, hypomethylated regions may also favor a permissive condition.
Indeed, the vast majority of genes induced at the time of ZGA or later
are either not significantly methylated, or hypomethylated, prior to
ZGA (
Andersen, Reiner, et al., 2012
). Additionally, hypomethylated regions