Biology Reference
In-Depth Information
with FeOOH (not AlOOH) under anaerobic (not oxygenated) conditions.
30,31
We hypothesized that these force-signatures originated from the unfolding
of cytochrome proteins, presumably to shuttled electrons to Fe(III), which
formed bonds between the bacterium and mineral.
0
0
100
100
200
200
300
300
400
400
500
500
600
600
0.0
0.0
0.0
0.0
-0.2
-0.2
-0.2
-0.2
-0.4
-0.4
-0.4
-0.4
-0.6
-0.6
-0.6
-0.6
green: Shewanella-AlOOH
blue: Shewanella-FeOOH
black: WLC 83 or 150 kD protein
green: Shewanella-AlOOH
blue: Shewanella-FeOOH
black: WLC 83 or 150 kD protein
-0.8
-0.8
-0.8
-0.8
-1.0
-1.0
-1.0
-1.0
0
0
100
100
200
200
300
300
400
400
500
500
600
600
distance (nm)
distance (nm)
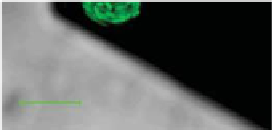
Figure 14.2.
(Left) Biologically active force probe showing living bacteria on the
end of an AFM cantilever.
14
Cells are luorescent green because of expression of an
intracellular green luorescent protein. Scale bar is ~10 μm. (Right) Force spectra for a
living
Shewanella oneidensis
bacterium on each of two minerals: goethite (FeOOH, light
and dark blue) and diaspore (AlOOH, light and dark green) immersed in an anaerobic
solution.
30,31
Black curves correspond to the modelled force-extension relationship for
two outer membrane proteins (83 and 150 kD) as determined by the worm-like chain
model.
14.4.3 Measuring Interacons Between Minerals and Pure
Proteins
We turned to the well-established worm-like chain (WLC) model to try to
determine whether the non-linear force-signatures observed in
Fig. 14.2
might be due to the unravelling of proteins that form a bond between the
bacterium and mineral surface. The WLC equation is as follows:
F
(
x
)
=
x
L
(
k
B
T
/
b
)
[0.25(1
/
L
)
2
+
x
/
0.25], where
F
is force (N),
x
is separation
or the extension distance of the protein (m),
k
B
is Boltzmann's constant
s
(1.381
10
23
J K
-1
),
T
is the temperature (298 K),
L
is the contour length of
the polypeptide of interest (in m) and
is its persistence length. For proteins,
the persistence length is often taken as the length scale of a single amino acid,
~0.4 nm.
b
By applying this equation to retraction proiles, one is able to
back out the contour length of the protein that forms the bond between two
surfaces. The size (in kD) of this protein can then be estimated by dividing the
contour length by 0.4 nm (the length scale of an individual amino acid) and
multiplying by 110 Da per amino acid.
47-49






























































































































Search WWH ::

Custom Search