Biology Reference
In-Depth Information
0
885,256
1,770,513
2,655,769
3,541,026
4,426,282
1,600,713
1,760,784
1,920,856
2,080,927
2,240,999
2,401,070
TR
b
RefSeq_genes
XenBase_genes
DR4
B
a
b
c
d
e
?
DNA-binding
Antibody
DNA
protein
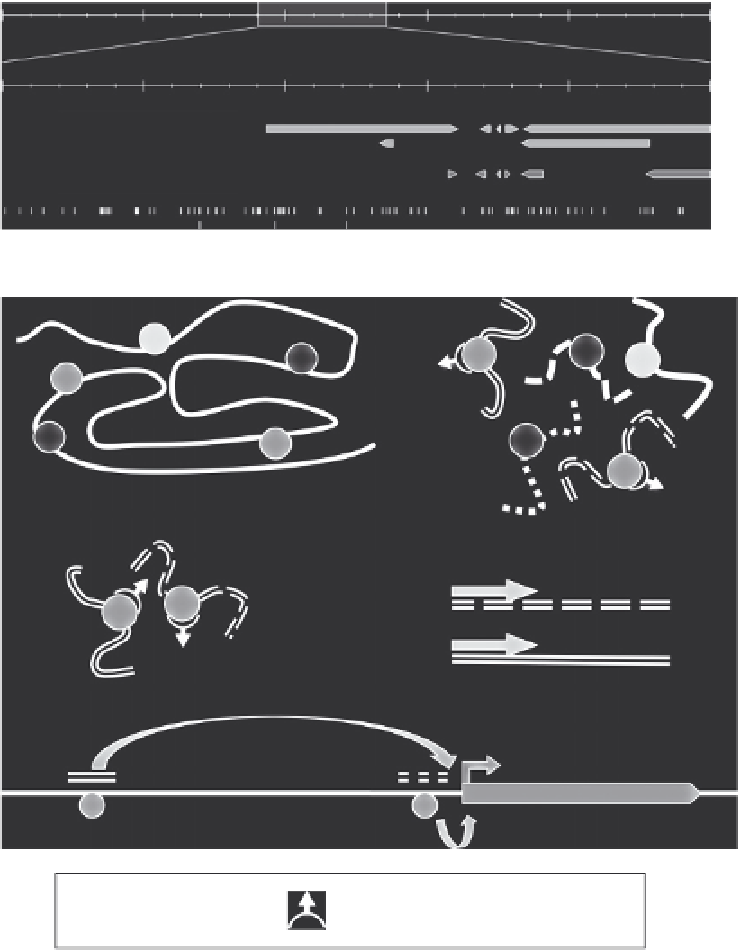
Figure 10.6 ChIP-Seq is a naive alternative to in silico predictions for mapping DNA-
binding sites. (A) Genome view of the TR
b
locus. The first two tracks represent the
XenBase and Ensembl gene models, respectively. The third track represents 21,424 pu-
tative DR4 found in this region with a modified version of NHRscan (
Sandelin &
Wasserman, 2005
). (B) Principle and limitations of ChIP-Seq. Physical interactions be-
tween chromatin components (protein, DNA) are stabilized by cross-linking (a). Chroma-
tin is then fragmented by sonication in fragments of 200
400 bp (b). The DNA-binding
sites are captured by immunoprecipitation with a protein-specific antibody (c). After
cross-link reversal, the DNA sample is purified and sequenced (d). Binding density pro-
files are derived from sequenced reads after mapping to the reference genome, and
provide an accurate description of the binding sites (e). Transcription factors (TFs) reg-
ulate the transcription of target genes after binding to DNA at enhancers, which can be
located far from genes. Unfortunately, ChIP-Seq cannot connect DNA-binding sites of
TFs to their target gene and thus cannot be used to describe the topology of gene reg-
ulatory networks.
-