Biology Reference
In-Depth Information
A
b
a
d
c
f
e
B
TSS 1
TSS 2
TTS 1
TTS 2
1
2
3
4
C
Current
RNA-PET
1
3
4
2
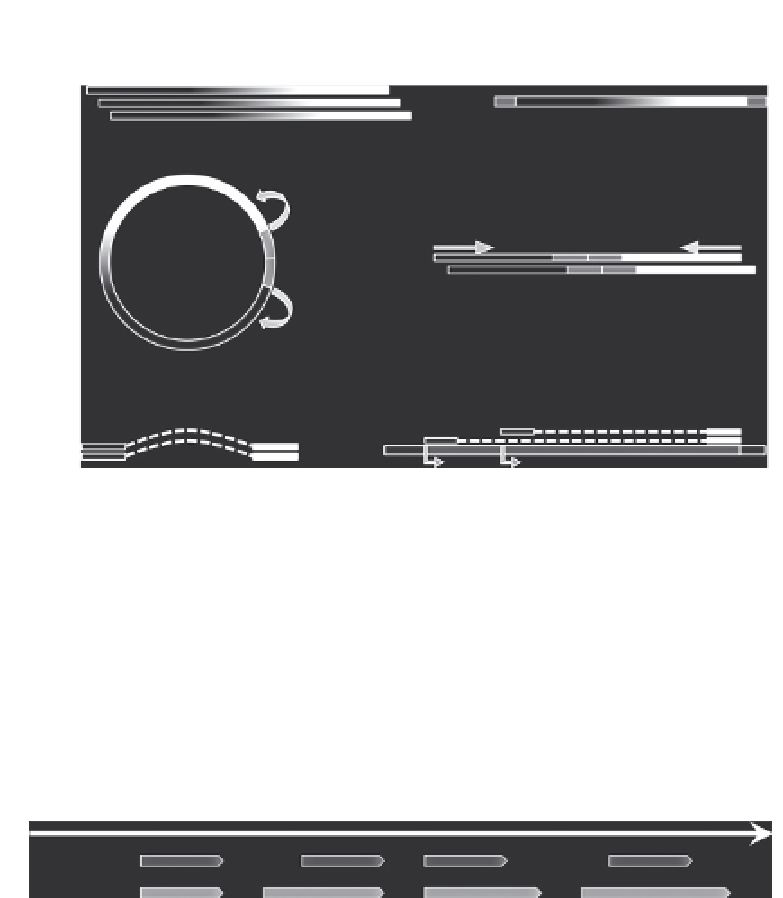
Figure 10.4 Principle of RNA-PET. (A) The starting point of RNA-PET is a library of full-
length cDNAs made from poly-A RNA (a), on which linkers containing Mme1 (or EcoP15I)
restriction sites are ligated (b). The resulting molecules are circularized and restricted
with
1 (c), which cuts DNA 27 bp downstream of its binding site and thus produces
a linear fragment containing the two ends of the original transcript separated by the
linker (d) prior to deep sequencing (from each end). The resulting paired-end tags (PETs)
contain information about the association of one transcript extremity to the other
Mme
—
respectively, white and black rectangles—(e). After mapping on the reference genome,
this provides experimental evidence for the location of the 5
0
and 3
0
ends of transcripts (f).
(B) Since RNA-PET captures the 5
0
and 3
0
ends of transcripts, it can be used to measure the
relative abundance of transcripts that use alternative TSS or TTS (dark segments on the full
gene model). (C) RNA-PET can improve the annotation of transcription units and thus
gene boundaries. From left to right, RNA-PET determined gene boundaries (dark gray)
can concord (1) with the current annotation (light gray), or extend in 5
0
(2), 3
0
(3), or both
(4) extremities.