Biology Reference
In-Depth Information
RNA Pol
II
miRNA gene
Transcription
Drosha
complex
Primary miRNA
AAAAA
Processing
Exportin-5
Precursor
miRNA
Transport
Dicer
complex
Processing
Strand
selection
GW182
Target
recognition
Argonaute
Argonaute
AAAAA
Target mRNA
Mature miRNA
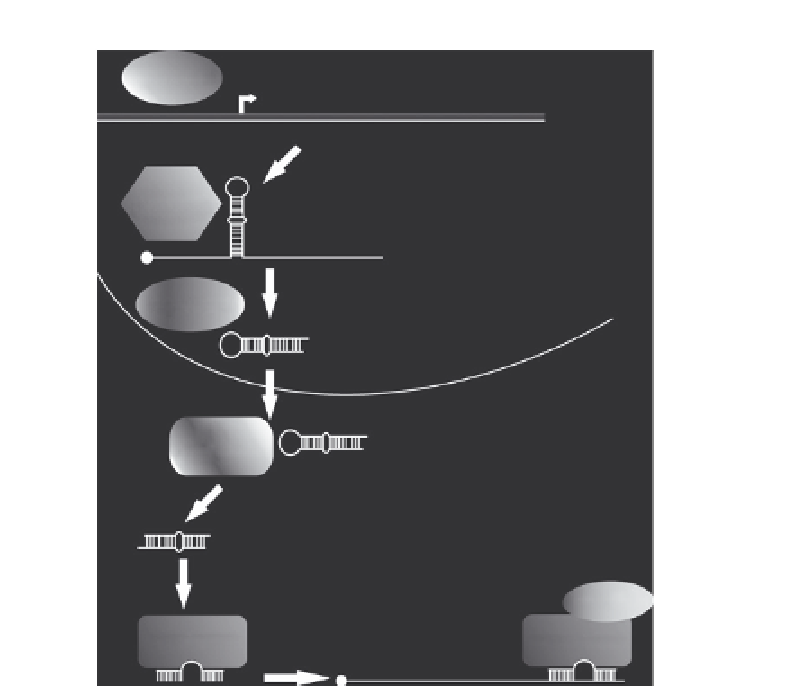
Figure 1.1
General miRNA biogenesis pathway. RNA polymerase II (RNA PolII)
transcribes miRNA genes to produce a capped and polyadenylated primary miRNA
(pri-miRNA). A complex containing the RNase III enzyme Drosha excises the
65nt
precursor miRNA (pre-miRNA), leaving a 2nt overhang at the 3
0
end. In flies and
vertebrates, nuclear transport protein Exportin-5, exports the pre-miRNA to the
cytoplasm, where it is subsequently cleaved by the RNase III enzyme Dicer to produce
an
21nt double-stranded product that features 2nt overhangs at both ends. The
mature miRNA strand is then loaded into Argonaute (Ago), where it works as part of
a complex with GW182, and other proteins to target mRNAs for repression.
common transcript (
Lau
et al.
, 2001; Lee and Ambros, 2001; Lim
et al.
,
2003
). The capped and polyadenylated transcripts, known as primary
miRNAs (pri-miRNAs), contain the stem-loop structure that houses the
functional
21nt miRNA sequence (
Kim
et al.
, 2009b
). The RNase III
enzyme Drosha, working in a complex with Pasha (also known as DGCR8),
excises the
65nt hairpin, forming what is known as the precursor miRNA
(pre-miRNA). A noncanonical Drosha-independent mechanism for produc-
ing pre-miRNAs was recently discovered in mammalian cells,
Drosophila
melanogaster
,and
Caenorhabditis elegans
(
Berezikov
et al.
, 2007; Okamura
et al.
, 2007; Ruby
et al.
,2007a
). In this pathway, pre-miRNAs derive from
debranching of short introns excised from pre-mRNAs. The spliced introns