Biology Reference
In-Depth Information
1.2. Signal flow and miRNAs
Signaling networks are crucial to establish expression patterns that lead to
cell decisions. In such systems, signal transduction has a directional flow; it
often begins with membrane-bound receptors that bind ligands to intracel-
lular transduction proteins interacting with other proteins, to translocation
of effectors to the nucleus resulting in altered gene expression.
Cui
et al
.
(2007)
found that miRNAs target signaling proteins of a human network
more frequently than what would be expected by chance. Importantly, the
distribution of miRNA targets in the signaling network is correlated with
signal flow and the position of the factors within the signaling network. The
propensity for a factor to be regulated by miRNAs increases in the direction
of signal flow, from ligands (9.1%) to cell surface receptors (18.8%),
to intracellular transducers (31.2%), and to nuclear proteins (50%) (
Cui
et al
., 2007
).
1.3. Network modules and miRNAs
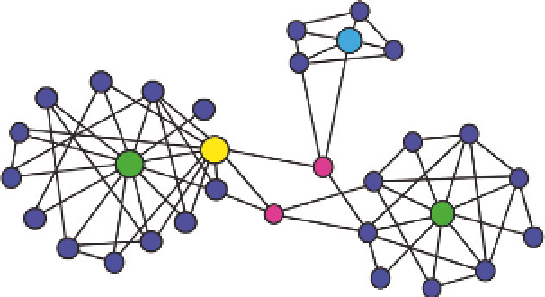
Biochemical networks are frequently organized into a set of distinct
subnetworks called modules. A module exists as a group of nodes that
are more highly connected to each other than to the rest of the network.
Modules are interconnected, typically through nodes called bottlenecks
(
Fig. 9.3
). Bottlenecks can in some cases be hubs. Since bottlenecks
connect modules, they have a high betweenness centrality, that is, are
nodes with many “shortest paths” going through them. They are analo-
gous to major bridges and tunnels with multiple parallel links between
Module C
Module A
Module B
Figure 9.3
Modular design of biochemical networks. A network with three modules.
Modules A and B have hubs (green) and bottlenecks (yellow and red). Module C has a
hub-bottleneck (blue).