Biology Reference
In-Depth Information
generate network robustness is to dampen, denoise, and set thresholds for
direct targets in a network. But as we shall see, miRNAs also generate
network robustness because of biases in the kinds of targets that they
regulate. Understanding how miRNAs do so will provide insight about
how higher-level biological processes such as development are also made
robust.
1.1. Network hubs and miRNAs
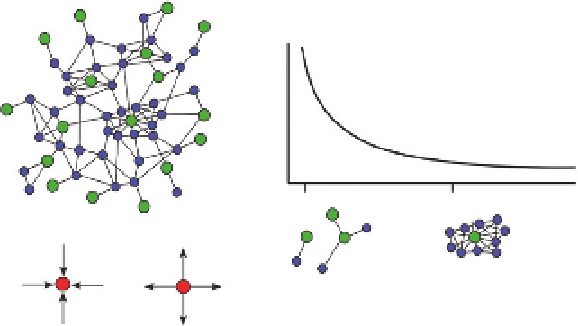
Molecular regulatory systems can be represented as networks composed of
nodes and links (
Fig. 9.1
A). Nodes can be genes, sequence elements, or
molecules such as proteins, metabolites, RNAs, etc. Links are the molecular
interactions between the nodes. The degree of a node is the number of links
that a node has with other nodes in the network. The collection of degrees
for each node in the network is the degree distribution and is frequently
represented as a graph of the frequency of each node degree type
(
Fig. 9.1
B). While some links such as protein-protein interactions do not
necessarily have an associated direction (undirected link), many links such as
between a transcription factor (TF) and its gene target, or a miRNA and its
mRNA target, are directed (i.e., a miRNA represses a target mRNA when
the two are bound together, and not the other way around). In directed
A
B
1
Degree
10
C
In degree
Out degree
Figure 9.1
Biochemical network organization. (A) Schematized network of nodes
(circles) and links (lines). Different molecular classes of nodes are highlighted in green
and blue. (B) A typical degree distribution for a network, illustrating that most nodes
have few links and a few nodes have many links. This organization gives rise to a power
law distribution that has a long tail. (C) Each link between nodes can be directed in
terms of cause-effect relationship. Links directed into a node effect that node, while
links directed out from a node effect the other node connected by the link.