Biomedical Engineering Reference
In-Depth Information
5.4
Positioning Digestion of Single DNA Molecules
with Nonspecific Endonuclease
Controlling chemical and biochemical reactions at nanometer scale has attracted
substantial attention from chemical and biological societies because of its potential
to explore new phenomena and/or unravel novel mechanisms that are inaccessible
in traditional bulk systems [
22
]. However, in the case of above-mentioned AFM
manipulation of DNA molecules, DNA was cut by the mechanical force applied by
the AFM tip. Under such conditions, the information about which bond on the DNA
backbone was broken remains unclear.
Herein, we introduce that a positioning scission reaction on a single DNA
molecule can be realized by using dip-pen nanolithography (DPN) method [
23
]. The
conventional DPN [
24
] is an AFM-based patterning technique in which molecules
are first attached to AFM tip and then transported to a solid surface when the AFM
tip was in contact with the surface. By employing the DPN technique, enzyme
molecules can be deposited onto selected positions on a DNA, so that positioning
scission reaction on a single DNA molecule can be realized (Fig.
5.6
). Since the
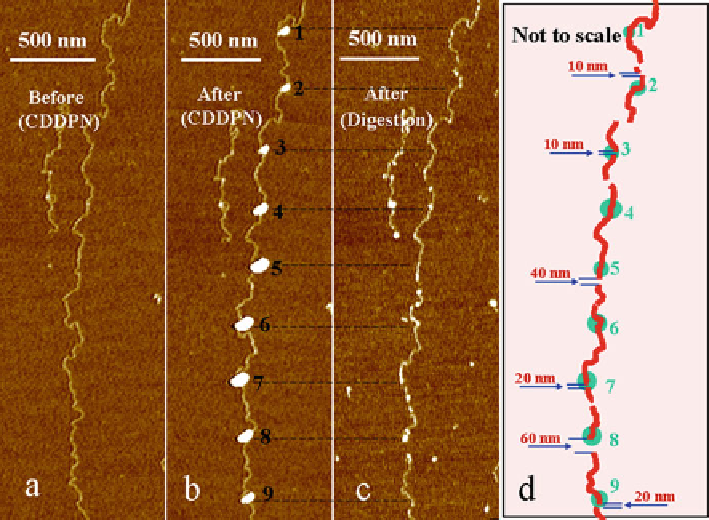
Fig. 5.6
DNA
molecule by DNase I. (
a
) Original DNA molecule. (
b
) Dcrops of DNase I on DNA molecule
after DPN deposition. (
c
) DNA fragments after digestion at desired sites by DNase I. (
d
) A sketch
indicating the digested sites on the DNA molecule. The
circles
represent the enzyme spots, and the
lines
represent the resulting DNA fragments after digestion (Reprinted with the permission from
Ref. [
23
]. Copyright 2007 American Chemical Society)
AFM images and a sketch indicating positioning digestion of an individual
œ
Search WWH ::

Custom Search