Biomedical Engineering Reference
In-Depth Information
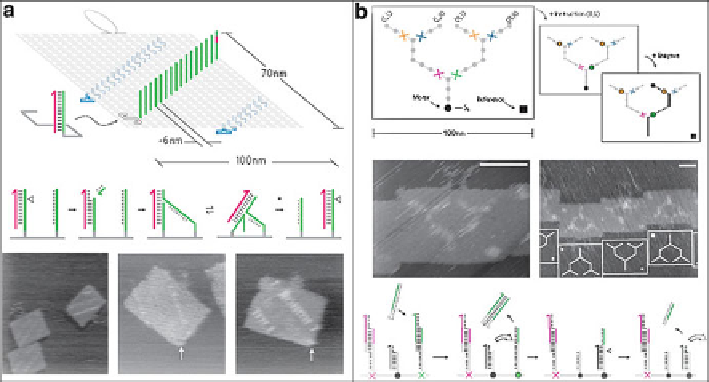
Fig. 12.6
(
a
) DNA motor and track (Reprinted by permission from Macmillan Publishers Ltd:
Ref. [
26
], copyright 2011). (
b
) Programmed route on branching tracks (Reprinted by permission
from Macmillan Publishers Ltd: Ref. [
27
], copyright 2012)
move on (Fig.
12.5
b). As the spider moved forward, the trailing track was degraded,
resulting in unidirectional and completely autonomous movement. This movement
was monitored by AFM and super-resolution total-internal-reflection fluorescence
microscopy. These two systems made people see the walker's moving under AFM
at the first time.
Just like camera taking photos, AFM only captured the moment of walker's
moving, which was not a real tracing. After the invention and development of high-
speed AFM, the walker's moving could be tracked. Sugiyama, Turberfield, and
coworkers reported a DNA motor moved autonomously and at a constant average
speed along the full length of the track on the origami [
26
]. It was a journey
comprising 16 consecutive steps for the motor which was monitored by high-speed
AFM (Fig.
12.6
a). The average motor speed is 0.1 nm
s
1
, which quantitatively
agreed with the data gotten from the simple model for stepping using first-order
rate constants obtained from experiments on a short track. Several months later,
they reported a similar enzyme-driving walker, the special point of which lied on
stator design. Two special control stators are positioned directly downstream from
the node (Fig.
12.6
b). Unique address and toehold sequences allow these control
stators to be selectively blocked and unblocked to direct a motor down a particular
path [
27
]. Also the progress was monitored by high-speed AFM and fluorescence
measurement.
Search WWH ::

Custom Search