Biomedical Engineering Reference
In-Depth Information
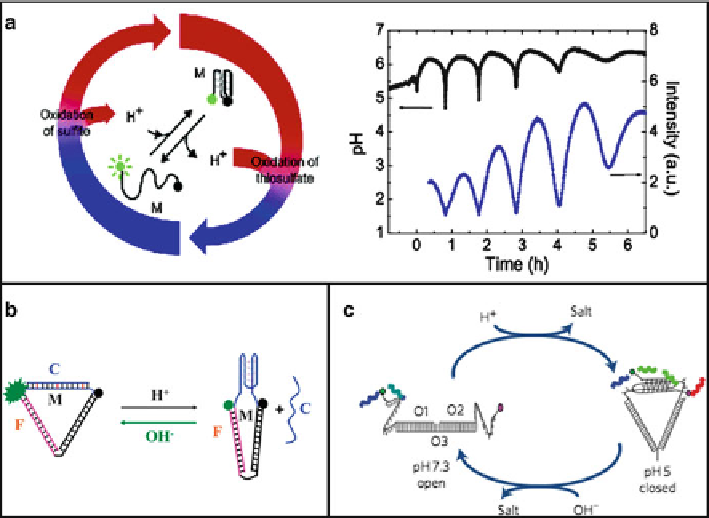
Fig. 11.4
(
a
) Operation cycle for DNA switch M driven by a chemical oscillator. An oscillatory
variant of the Landolt reaction changes the pH value of the reaction solution periodically. In one-
half of the reaction cycle, protons produced during the oxidation of sulfite induce a conformational
transition to a folded DNA structure, the so-called i-motif. In the other half of the reaction cycle, the
oxidation of thiosulfate consumes protons and leads to an unfolding of the i-motif (Reprinted with
permission from Ref. [
33
]. Copyright 2005 American Chemical Society). (
b
)and(
c
) i-motif-based
switches inducing second-order lever strand motions (Reproduced from Ref. [
37
] by permission
of The Royal Society of Chemistry. Reprinted by permission from Macmillan Publishers Ltd: Ref.
[
38
], copyright 2009)
cation recombined the hydroxide ions and returned to MGCB, resulting in the
acidic solution and i-motif conformation. Furthermore, an oscillatory variant of the
Landolt reaction that changes pH value of the reaction solution periodically was
utilized to induce the folding and unfolding of i-motif structure [
33
](Fig.
11.4
a).
The switch rate of DNA structure tracked by FRET was in high agreement with the
pH oscillation rate monitored with pH meter. More observable ways are exploited
to track the mechanical motion of i-motif-based switches by utilization of the
assembly properties of DNA-functionalized gold nanoparticles (Au NPs). The Au
NPs coated with C-rich strands were isolated in high-pH solution showing red
in color because of the electrostatic repulsion between negatively charged DNA
strands taking random coil conformation. While in low-pH solution, the formation
of interparticle i-motif structures brought the Au NPs together to form aggregates,
which showed purple in color [
34
]. In another way, C-rich strands coating on Au
NPs folded into intraparticle i-motif structures in low-pH solution while stretched
Search WWH ::

Custom Search