Biomedical Engineering Reference
In-Depth Information
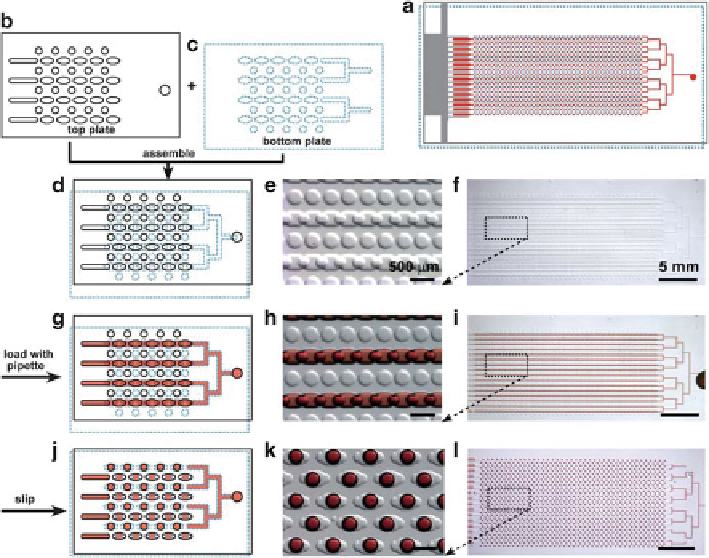
Fig. 7.16
Digital PCR on SlipChip. (
a
) The entire SlipChip is assembled by two plates: The
top
plate
is outlined with a
black solid line
(
b
), the
bottom plate
is outlined with a
blue dotted line
(
c
), and
red
represents the sample; (
d
-
f
) the SlipChip is assembled such that the elongated wells
in the
top and bottom plates
overlapped to form a continuous fluidic path; (
g
-
i
) the aqueous
reagent (
red
) was injected into SlipChip and filled the chip through the connected elongated
wells; (
j
-
l
)the
bottom plate
was slipped relative to the
top plate
such that the fluidic path was
broken up and the
circular wells
were overlaid with the
elongated wells
, and aqueous droplets
were formed in each compartment. (
d
), (
g
), and (
j
): schematic of the SlipChip; (
e
), (
h
), and
(
k
): zoomed in microphotograph of the SlipChip; (
f
), (
i
), and (
l
): microphotograph of the entire
SlipChip (Reproduced from Ref. [
72
] with permission of The Royal Society of Chemistry)
using a minimal number of wells. The smallest wells enable quantification of
high concentrations, while the wells of large volumes enable high sensitivity by
efficiently increasing the total volume. They realized the design on the SlipChip
system (Fig.
7.17
), and the chip allows for simpler device design and minimizes
reagent consumption.
Digital PCR can also be realized by emulsion droplet techniques, another com-
partmentalization strategy which has been discussed in the previous Sect.
7.2.1.2
.
Hindson and coworkers demonstrated a flow-focusing device that enables a high-
throughput digital PCR amplification using conventional TaqMan assays [
84
]. The
uniform droplets were efficiently generated and collected into a microtiter plate and
thermal cycled to the endpoint. After thermal cycling, the droplets from each well
were aspirated and streamed toward a detector where a spacer fluid separated and
aligned them for single-file fast detection (Fig.
7.18
).
Search WWH ::

Custom Search