Biology Reference
In-Depth Information
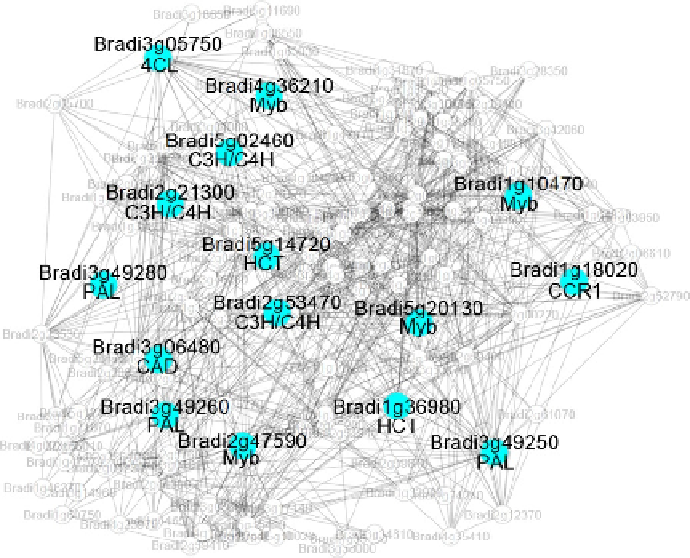
Fig. 3. Co-expression network surrounding PAL (Bradi3g49260) from Brachypo-
dium distachyon. Nodes and edges represent genes and significant positive co-expression
relationships between genes. Blue nodes depict genes with known association to lignin
biosynthesis while grey nodes represent genes with no known involvement in lignin
biosynthesis. Abbreviations below the gene identifiers correspond to MYB (MYB
transcription factor family), PAL (phenylalanine ammonia-lyase), C3H/C4H (p-cou-
marate 3-hydroxylase/trans-cinnamate 4-hydroxylase), HCT (hydroxycinnamoyl trans-
ferase), CAD (cinnamyl alcohol dehydrogenase) and 4CL (4-coumarate: CoA ligase).
enrichment in genes involved in lignin biosynthesis (p-value
0.01, data now
shown), further supporting involvement of the genes in lignin biosynthesis.
Interestingly, the network is also significantly associated with phenylalanine
biosynthesis, indicating that production of phenylalanine is coupled with
biosynthesis of lignin. In addition, genes involved in cellulose biosynthesis
(such as cellulose synthases) are also significantly represented, suggesting
coordination of cellulose biosynthesis and lignin deposition. Finally, the net-
work contains many uncharacterized genes that may potentially be involved in
lignin biosynthesis as well.
However, it is important to keep in mind caveats inherent with co-
expression analysis: (i) Representation of genomic content on the microarrays
<