Biology Reference
In-Depth Information
No crystal structure is available for cyanobacterial Fur homologues,
though their main folding is probably shared with other Fur proteins. In
fact, FurA from
Anabaena
PCC 7120 shows around 40% sequence similar-
ity with
Pseudomonas
Fur and has similar helical content, as demonstrated
by FTIR and far-UV CD (
Hernández et al., 2005
). A three-dimensional
model for the FurA monomer from
Anabaena
PCC 7120 has been
obtained by homology modelling based on its similarity with
P. aeruginosa
Fur, although the lack of strong sequence identity at the dimerization
interface between
P. aeruginosa
Fur and FurA precluded dimer modelling
(
Hernández et al., 2005
). Homology modelling has also been used to build
the 3D structure of the PerR-like regulator Slr1738 from
Synechocystis
PCC 6803 (37% homology with
P. aeruginosa
Fur protein) and its target
DNA (
Garcin et al., 2012
).
3.1.1.1. Metal-binding site
Fur proteins show two potential metal-binding motifs rich in histidines
and cysteines; a conserved HHXHX
2
CX
2
C and another, less conserved,
carboxyl-terminal motif CX
2
C (
Fig. 4.2
). The structure of Fur from
P. aeruginosa
exhibits two metal-binding sites (
Pohl et al., 2003
). Site 1,
placed in the dimerization domain, comprises the side chain of residues
His
86
, Asp
88
, Glu
107
and His
124
and a water molecule resulting in a distorted
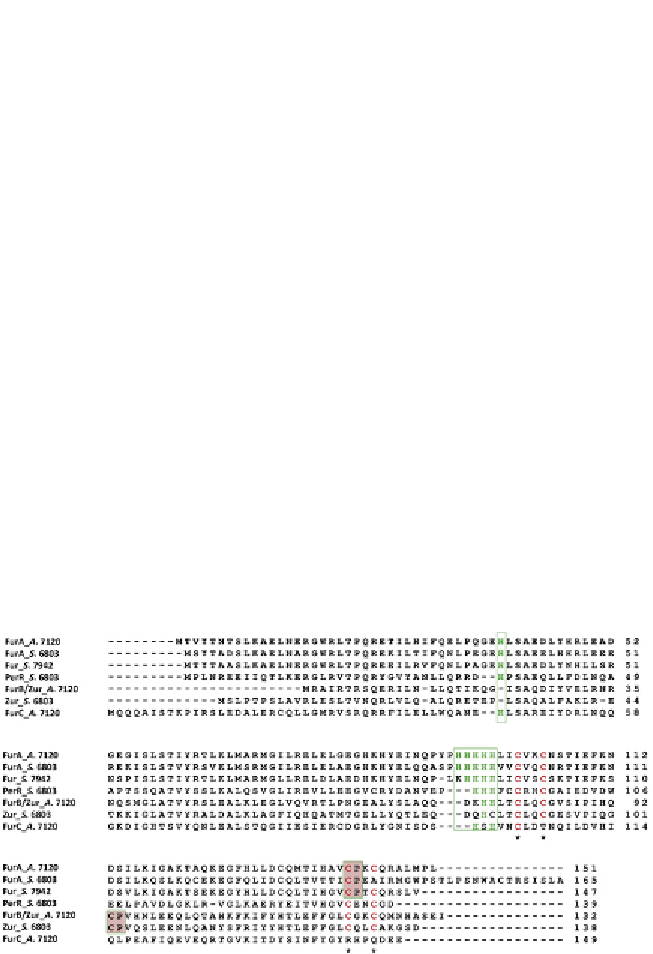
Figure 4.2
Alignment of a representative subset of different members of the Fur
family from cyanobacteria. A. 7120,
Anabaena
PCC 7120; S. 6803,
Synechocystis
PCC
6803; S. 7942,
Synechococcus
PCC 7942. The conserved histidine in the N-terminal
domain potentially involved in DNA-binding and the His-rich motif are boxed. Cysteine
residues in the CXXC redox motifs are indicated with asterisks. Haeme-regulatory CP
motifs are indicated in grey boxes. For colour version of this figure, the reader is referred
to the online version of this topic.