Biomedical Engineering Reference
In-Depth Information
(A)
(B)
E
L
O
N
G
A
T
I
O
N
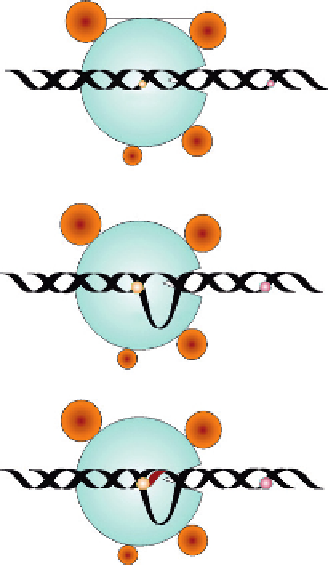
RNA polymerase
Star site
Stop site
DNA-RNA hybrid
Nascent RNA
I
N
I
T
I
A
T
I
O
N
(C)
T
E
R
M
I
N
A
T
I
O
N
Transcription bubble
Completed
RNA strand
Figure 1.6
Steps in transcription: (A) initiation; (B) elongation; and (C) termination.
an operon. The transcription of operon produces polycistronic mRNA that codes for
several functionally related proteins. Such clustering of genes is not seen in eukar-
yotes, and in fact the genes for related proteins are often present on different chro-
mosomes and transcribed separately. But the eukaryotic genes have parts of coding
sequences (exons) intervened by noncoding sequences (introns), requiring that the
long mRNA strand transcribed be spliced or clipped to remove the noncoding parts
and then ligated back together. Following the above modifications, the pre-mRNA
transcript containing exons and introns is spliced to form functional mRNA. This is
accomplished by spliceosome, wherein a group of proteins, along with small nuclear
RNA, splice together the exons by forming loops of the introns and excising them.
In contrast, prokaryotes can exhibit transcription and translation simultaneously,
as generally noncoding sequences are nonexistent in such cells. Moreover, there is
no separate nuclear and cytoplasmic region to separate the two processes. However,
in eukaryotes, mRNA primary transcripts must undergo processing in the nucleus
before they are functional in the cytoplasm. As the primary RNA transcript dissoc-
iates from the RNA polymerase, a 5 cap is formed by the addition of methyl guany-
late at the 5 end. This cap allows the easy export of RNA to cytoplasm and is the
site for a specific protein association that is required to initiate translation. At the

Search WWH ::

Custom Search