Image Processing Reference
In-Depth Information
Spatial smoothing
Spatial smoothing
13
13
−
8.00
−
8.00
3.20
3.20
−
−
8.00
8.00
3.20
t (123)
p < 0.001749
3.20
t (123)
p < 0.001749
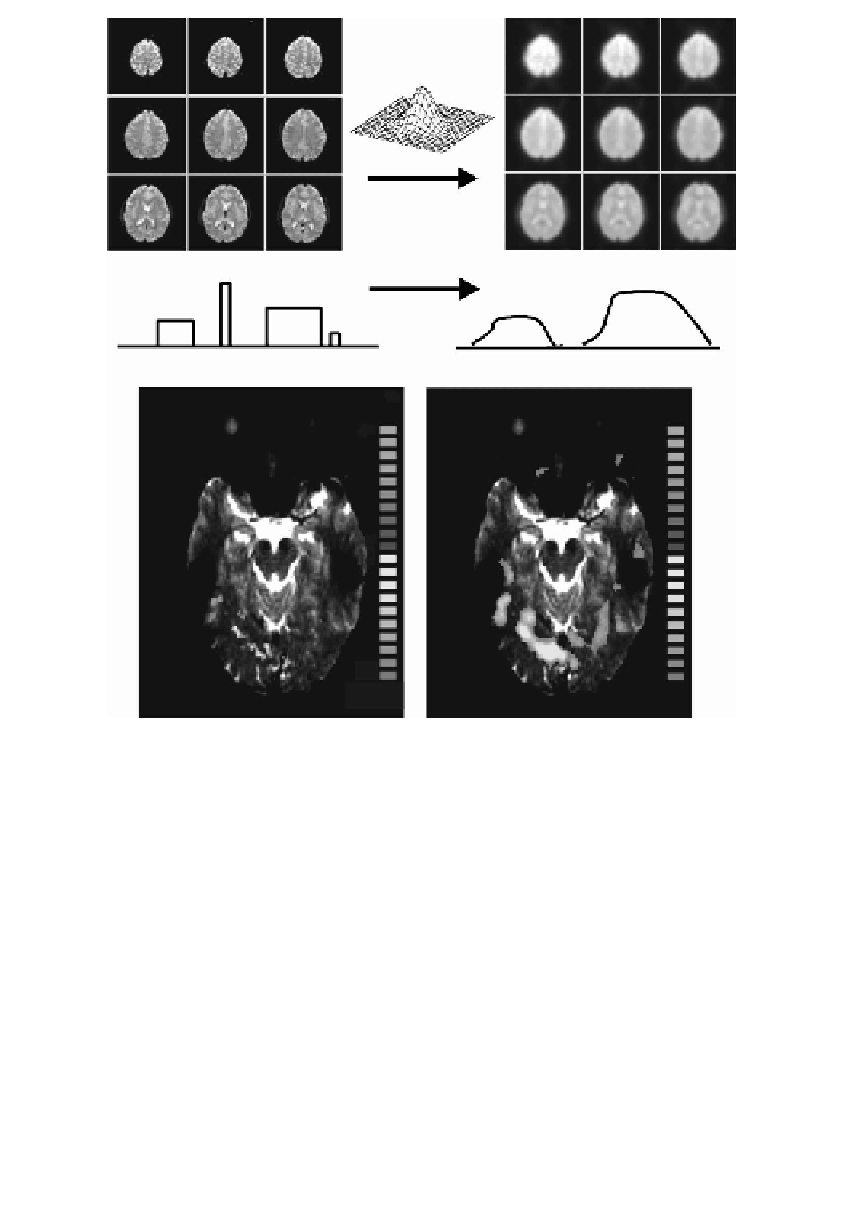
FIGURE 15.3
Spatial smoothing. Top row: Functional time series before (left) and after
(right) spatial filtering with a 3-D Gaussian smoothing kernel (FWHM = 3 voxels). Middle
row: Schematic representation of the effects of spatial smoothing on brain activation. For
simplicity, space is represented on a 1-D axis. Note that “activated” regions with a small
spatial extension may disappear after spatial smoothing. Lower row: Activation map
before (left) and after (right) spatial smoothing with a 3-D Gaussian smoothing kernel
(FWHM = 3 voxels).
MR signal drifts and physiological fluctuations at low temporal frequencies
are always present in fMRI time series and have to be corrected by linear
detrending or, more generally, by high-pass temporal filtering (see
Figure 15.4
)
[18-21]. If the temporal sampling rate used for functional scanning (volume
repetition time) is sufficiently short, systematic physiological noise linked to the
cardiac and respiratory cycles can be eliminated using band-reject [20] or least-
mean-square (LMS) adaptive filters [21].





































Search WWH ::

Custom Search