Database Reference
In-Depth Information
2.
Now we can get the indexes of the data rows that are in each cluster by looking at
the
:sets
key of
iris-clusters
. Then we can pull the species from each cluster's
rows to look at the frequency of each species in each cluster:
user=> (doseq [[pos rws] (:sets iris-clusters)]
(println pos \:
(frequencies
(i/sel iris :cols :Species
:rows rws))))
[4 1] : {virginica 23}
[8 1] : {virginica 27, versicolor 50}
[9 0] : {setosa 50}
So we can see that
setosa
and
versicolor
are each put into their own clusters, and half of
the virginica are in their own cluster and half are with the versicolors.
How it works…
SOMs use a neural network to map data points onto a grid. As the neural network is trained,
the data points converge into cells in the grid, based on the similarities between the items.
We can get the size of the output map using the
:dims
key:
user=> (:dims iris-clusters)
[10.0 2.0]
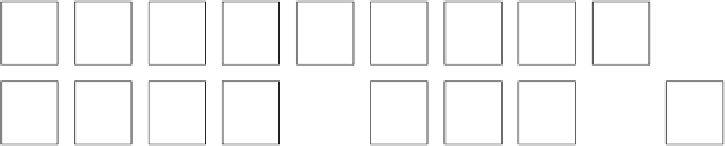
We can use this information, combined with the cell frequencies, to graph the clustering of
data in the SOM:
50
0
0
50
0
0
50
0
0
Virginica
Setosa
Versicolor
One of the downsides of SOMs is that the network's weights are largely opaque. We can
see the groupings, but iguring out why the algorithm grouped them the way it did is dificult
to deine.