Biology Reference
In-Depth Information
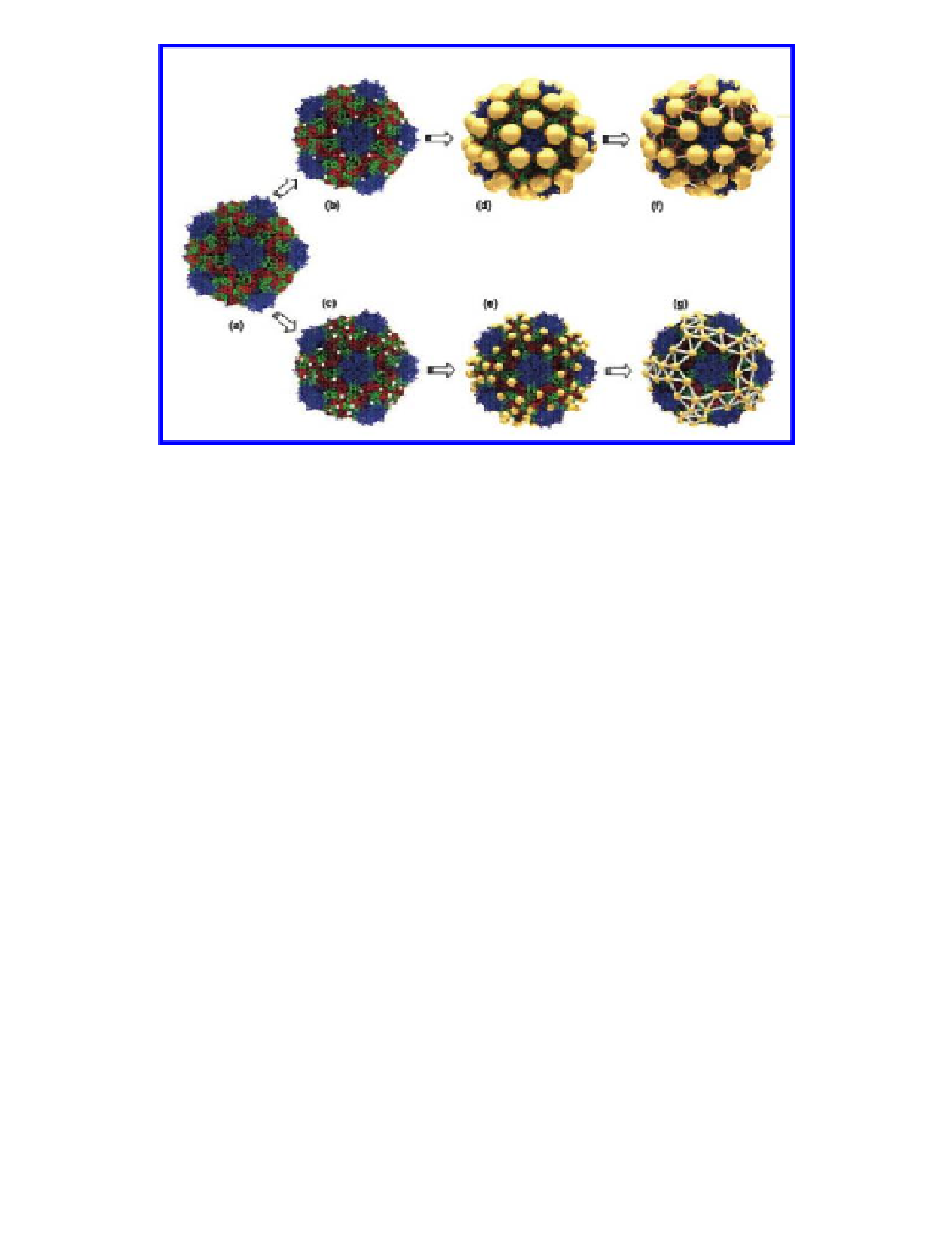
Figure 4.19
Schematic of the procedure used to create molecular networks on the
surface of the virus capsid: (A) CPMV capsid structure from crystallographic data;
(B) mutant with one cysteine (white dots) per subunit. The four nearest-neighbor
cysteine-to-cysteine distances are 5.3, 6.6, 7.5, and 7.9 nm; (C) double mutant with
two cysteines per subunit. The four nearest-neighbor cysteine-to-cysteine distances
are 3.2, 4.0, 4.0, and 4.2 nm; (D) mutant with 5-nm gold nanoparticles bound to the
inserted cysteines; (E) double mutant with 2-nm gold nanoparticles bound to the
inserted cysteines; (F) mutant with the 5-nm gold particles interconnected using
1,4-C
(red) and oligophenylenevinylene
(silver) molecules; (G) double mutant with the 2-nm gold particles interconnected
with oligophenylenevinylene molecules. Reproduced with permission from Blum, A.
S., Soto, C. M., Wilson, C. D., Brower, T. L., Pollack, S. K., Schull, T. L., Chatterji, A., Lin,
T., Johnson, J. E., Amsinck C., Franzon P, Shashidhar, R., and Ratna, B. R., (2005) An
engineered virus as a scaffold for three-dimensional self-assembly on the nanoscale,
Small
H
[
trans
-(
-AcSC
H
C
≡
CPt-(PBu
)
C
≡
C]
6
4
4
6
4
3
2
2
,
1
(7), 702-706.
.. Fluorescent-labeled VnPs as Signal enhancers in Biological
Assays
Biological assays such as DNA microchip arrays and immunoassays make use
of fluorescent-labeled molecules for detection. Because of the multivalent
nature of VNPs, multiple fluorescent dyes can be displayed on a single VNP,
thus enhancing signal sensitivity.
In a first example, VNP sensors were utilized in a DNA microarray
used for pathogen genotyping. Herein, CPMV particles were covalently
modified with approximately 40 dyes. Neutravidin (a protein that binds
biotin) was also attached to the VNPs and served as a recognition element.
Search WWH ::

Custom Search