Biology Reference
In-Depth Information
The structure of HIV-1 SL2, which binds specifically to the NC
protein, was determined by NMR spectroscopy.
70
The molecule adopts
a stem-loop structure in which the loop region is largely disordered.
The stem adopts a base pairing pattern that differs from the predicted
secondary structure. Instead of the predicted A:U base and a bulged
out A nucleotide, the structure contains an A-U-A base triple plat-
form (Fig. 7).
SL3 also adopts the expected stem-loop arrangement. The stem is a
Watson-Crick paired A-form helix and the loop is flexible, yet shows
some preferred stacking patterns for the bases at the loop-stem interface.
The structure of a complete
-site RNA was recently determined
for MMLV.
71
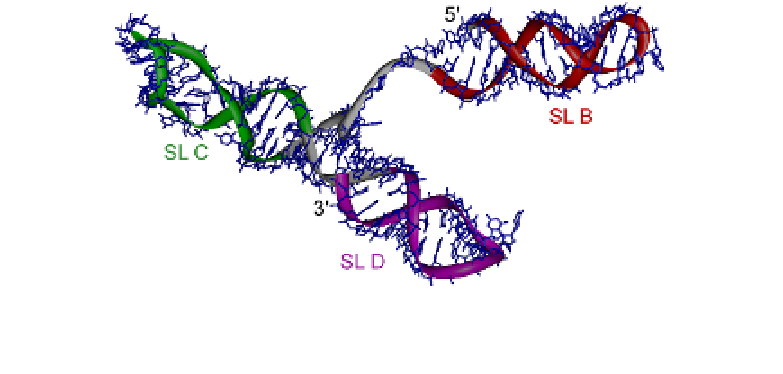
In this 101-nucleotide construct the three individual
stem-loops (SL-B, SL-C, and SL-D) of the core encapsidation signal
are well defined. SL-C and SL-D are stacking end-to-end on each
other and SL-B is connected to this longer segment via a conserved
flexible linker (Fig. 8).
Ψ
Complexes between the HIV-1
Ψ
-site RNAs and the NC
Protein
-site clearly have over-
lapping and possibly redundant functions. SL1 has its core function in
controlling the packaging of dimeric RNA. SL2 and SL3 can both
The four stem-loops that are part of the HIV-1
Ψ
Fig.8.
Structure of the core encapsidation signal RNA of MMLV (1S9S).