Information Technology Reference
In-Depth Information
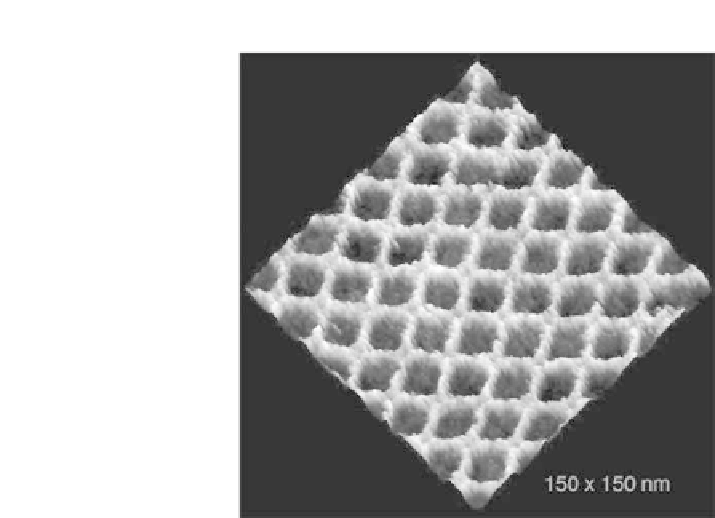
Figure
13.5.
AFM image of 2D lattice of cross tiles.
a bit-wise cumulative XOR computation. (Given n bits as input, each ith output is
the XOR of the first i input bits, which is the computation occurring when one
determines the output bits of a full-carry binary adder circuit.) The experiment [9]
is described further in Figure 13.6.
Figure 13.6 shows a unit TX tile (a) and the sets of input and output tiles
(b) with geometric shapes conveying sticky-end complementary matching. The
input layer and corner condition tiles were designed to assemble first [see example
computational assemblies in Fig. 13.6 (c) and (d)]. The output layer would then
assemble specifically starting from the bottom left using the inputs from the input
layer. The tiles were designed such that an output reporter strand ran through all
the n tiles of the assembly by bridges across the adjoining pads in input, corner,
and output tiles. This reporter strand was pasted together from the short ssDNA
sequences within the tiles using ligation enzyme mentioned previously. When the
solution was warmed, this output strand was isolated and identified. The output
data was read by experimentally determining the sequence of cut sites (as
described below). In principle, the purified output strands could be used for
subsequent computations.
This experiment [9] provided answers to a basic question:
How can one provide data input to a molecular computation using DNA tiles?
In this experiment the input bits (1's and 0's) were encoded on two different
tile types with specific sticky-ends and specific endonuclease cleavage sites





Search WWH ::

Custom Search