Information Technology Reference
In-Depth Information
formal model of 2D tiling due to Wang (in 1961), which culminated in a proof by
Berger in 1966 that universal computation could be done via tiling assemblies.
Winfree [7] was the first to apply the concepts of computational tiling assemblies
to DNA molecular constructs. His core idea was to use tiles composed of DNA to
perform computations during the process of self-assembly, where only valid
solutions to the computation are allowed to assemble. To understand this idea,
we will need an overview of DNA nanostructures, as presented in the next section.
13.4.2. DNA Nanostructures
Recall that a DNA nanostructure is a multimolecular complex consisting of a
number of ssDNA that have partially hybridized along their subsegments. The
field of DNA nanostructures was pioneered by Seeman [4].
Particularly useful types of motifs often found in DNA nanostructures include
stem-loops and sticky ends, as illustrated below.
Figure 13.2a illustrates a stem-loop where ssDNA loops back to hybridize on
itself; that is, one segment of the ssDNA (near the 5
u
end) hybridizes with another
segment further along (nearer the 3
end) on the same ssDNA strand. The shown
stem consists of the dsDNA region with sequence CACGGTGC on the bottom
u
3
′
A
C
T
G
T
G
C
C
A
C
G
T
T
C
A
C
GG
T
G
C
T
A
C
(a)
5
′
3
′
5
′
T
G
C
G
G
C
C
A
A
T
C
G
3
′
5
′
(b)
Figure
13.2.
A stem-loop and a sticky end.




















































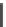

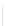




Search WWH ::

Custom Search