Information Technology Reference
In-Depth Information
Molecular-scale devices using DNA nanostructures have been engineered to
have various capabilities, ranging from (i) the execution of molecular-scale computa-
tion, (ii) use as scaffolds or templates for the further assembly of other materials
(such as scaffolds for various hybrid molecular electronic architectures or perhaps
high-efficiency solar-cells), (iii) robotic movement and molecular transport, and
(iv) exquisitely sensitive molecular detection and amplification of single molecular
events, and (v) transduction of molecular sensing to provide drug delivery.
13.2. INTRODUCTORY DEFINITIONS
13.2.1. A Brief Introduction to DNA
Single stranded DNA (denoted ssDNA) is a linear polymer consisting of a
sequence of DNA bases oriented along a backbone with chemical directionality.
By convention, the base sequence is listed starting from the 5-prime end of the
polymer and ending at the 3-prime end (these names refer to particular carbon
atoms in the deoxyribose sugar units of the sugar-phosphate backbone, the details
of which are not critical to the present discussion). The consecutive nucleotide
bases (monomer units) of an ssDNA molecule are joined through the backbone
via covalent bonds. There are four types of DNA bases: adenine, thymine,
guanine, and cytosine, typically denoted by the symbols A, T, G, and C,
respectively. These bases form the alphabet of DNA; the specific base sequence
comprises DNA's information content. The bases are grouped into complementary
pairs (G, C) and (A, T).
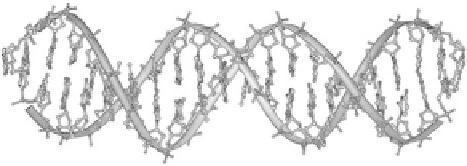
The most basic DNA operation is hybridization, where two ssDNA oriented in
opposite directions can bind to form a double stranded DNA helix (dsDNA) by
pairing between complementary bases. DNA hybridization occurs in a buffer
solution with appropriate temperature, pH, and salinity. A dsDNA helix is
illustrated in Figure 13.1.
Since the binding energy of the pair (G, C) is approximately half-against the
binding energy of the pair (A, T), the association strength of hybridization
depends on the sequence of complementary bases, and can be approximated by
Figure
13.1.
Structure of a DNA double helix (Created by Michael Str¨ ck and
released under the GNU Free Documentation License (GFDL).)






Search WWH ::

Custom Search