Biology Reference
In-Depth Information
or chlorine-related oxidants yielded 5-chloro-CYN and cylindrospermic acid and both of these
derivatives proved to be non-toxic even at doses 50 times higher (10 mg kg
-1
mouse body weight)
than LD
50
of CYN. These results emphasize that the pyrimidine ring of uracil is essential for toxicity
of CYN (Banker
et al
., 2000, 2001).
ii) Biosynthesis
:
The biosynthetic pathway of CYN suggests that glycine is the precursor and later
guanidinoacetic acid serves as the starter unit for the polyketide chain. C1 and C2 of glycine serve
as C14 and C15 of CYN. Carbon atoms 4 to 13 arise from contiguous acetate units attached head
to tail. The origin of NH-CO-NH segment in the uracil ring is not known (Bourgoyne
et al
., 2000).
The involvement of three genes
aoaA
,
aoaB
and
aoaC
that encode amidinotransferase, a hybrid
NRPS/PKS and PKS, respectively have been reported to mediate the biosynthesis of CYN in
Aph.
ovalisporum
(Shalev-Alon
et al
., 2002). The synthesis of guanidinoacetic acid is mediated by the
enzyme amidinotransferase which is then passed on to the hybrid NRPS/PKS and PKS enzyme
systems for further synthesis. The characterization of amidinotransferase from
C
.
raciborskii
AWT205
has been reported (Kellmann
et al
., 2006). A
cyr
gene cluster (
cyrA
to
cyrO
) for biosynthesis of CYN
in
C
.
raciborskii
AWT205 has been sequenced and characterized. A 43 kb fragment of DNA consists
of 15 ORFs that mediate biosynthesis, regulation and export of the toxin. The fl anking regions on
either side of the
cyr
cluster consists of
hyp
genes, the gene products of which act as molecular
chaperones for the maturation step of hydrogenases. The precursor molecule glycine accepts an
amino group via an amidinotransfer reaction that constitutes the fi rst step. This is soon followed
by fi ve polyketide extensions and subsequent reduction reactions (Mihali
et al
., 2008). Stucken
et al
.
(2010) sequenced the genomes of
C
.
raciborskii
CS-505 and
Raphidiopsis brookii
D9. In the former the
cyr
gene cluster is spread over 41.6 kb region and organized into 16 ORFs that bear a high degree of
synteny with those of the genes in the
cyr
gene cluster of
C
.
raciborskii
AWT205. This resemblance
in the two gene clusters is not only limited to the arrangement of genes but also to the fl anking
regions in having
hyp
genes at both ends of the
cyr
gene cluster. In addition to
cyrL
and
cyrM
,
C
.
raciborskii
CS-505
cyr
gene cluster revealed a 219 bp transposase element that is located between
cyrC
and
cyrE
(Fig. 11). This particular region with the transposase elements is also refl ected in the
CYN biosynthesis gene cluster of
R
.
brookii
D9 indicating that rearrangements have occurred in this
section of the genome. The announcement of draft genome sequence of
Oscillatoria
PCC 6506 with
several gene clusters responsible for toxins and secondary metabolites (Méjean
et al
., 2010) and
subsequent identifi cation of complete gene cluster for CYN and 7-epi-CYN biosynthesis paved the
way for understanding the complete set of biosynthetic reactions (Mazmouz
et al
., 2010). Though
this gene cluster is homologous with the gene cluster of
C
.
raciborskii
AWT205 there exist some
differences in the number of genes and their arrangement. Instead of 15
cyr
genes as in
C
.
raciborskii
,
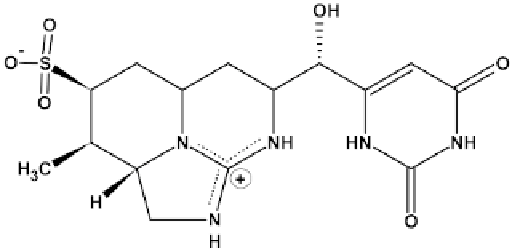
Figure 10:
Structure of CYN