Biology Reference
In-Depth Information
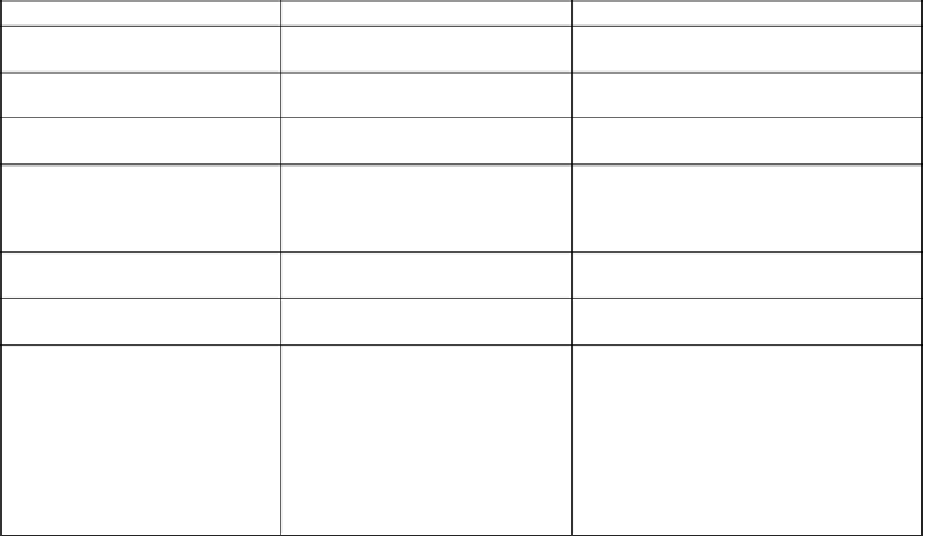
Table 9:
Prochlorococcus
cyanophages with their host of isolation and cross-infectivity between strains of the same genus and
those of
Synechococcus
* (after Sullivan
et al
., 2003).
Cyanophage(s)
Host on which isolated
Hosts on which cross-infection was possible
Syn1
Synechococcus
WH8101
Synechococcus
WH7803, WH8017 and
WH8018
Synechococcus
WH6501
S-SM1, S-ShM1 and S-SSM1
Synechococcus
WH8102
Syn9
Synechococcus
WH8109
Broad host range; infected HL-adapted MIT
9515 and MED4 strains of
Prochlorococcus
P-RSP3, P-SSP8
LL-adapted strain NATL2A of
Prochlorococcus
Did not cross infect any other strain of
Prochlorococcus
both HL-adapted as well
as LL-adapted strains and of
Synechococcus
strains
P-RSM3
LL-adapted strain NATL2A of
Prochlorococcus
Cross infected only LL-adapted MIT9211,
NATL1A of
Prochlorococcus
P-SSM3, P-SSM4, P-SSM5, P-SSM6,
P-RSM2
LL-adapted strain NATL2A of
Prochlorococcus
Cross infected other LL-adapted strains of
Prochlorococcus
P-RSP2
HL-adapted strain of
Prochlorococcus
MIT9302
HL-adapted strain of
Prochlorococcus
MIT9312
Did not cross infect other HL-adapted strains
or LL-adapted strains of
Prochlorococcus
or
other strains of marine
Synechococcus
strains
P-SSP2, P-SSP4
Cross-infected only
high-light adapted
strain of
Prochlorococcus
MIT9515
*
Synechococcus
strain WH8020 did not support the growth of any of the 44 phage strains tested (see text). HL-high light;
LL-low light.
P-SSP3
HL-adapted strain of
Prochlorococcus
MIT9312
SPGM-03) had a very restricted host range as they infected only one strain of the host whereas majority
of the cyanophage isolates (SPGM-04, SPGM-11 to SPGM-35) exhibited a broad host range as they
infected 10 to 17 strains of the host. The rest of the cyanophages (SPGM-05 to SPGM-10) infected
3 to 8 host strains and so had an intermediate host range (McDaniel
et al
., 2006). Virus isolation
studies conducted from Indian Ocean by plaque assay method on
Synechococcus
WH7803 revealed
that cyanophages were abundant from the surface waters to a depth of 10 m with a maximum at
1.00 am after which there was a decrease in their numbers. Nearly 50 such cyanophages (S-IO1 to
S-IO50) have been isolated whose morphotype could not be established (Clokie
et al
., 2006). Tables
6 and 7 summarize some of the marine cyanophages isolated and characterized against marine
Synechococcus
and
Prochlorococcus
strains. A probable explanation that is given for the cross-infectivity
of
Prochlorococcus
and
Synechococcus
strains by the marine cyanomyoviruses is the existence of
additional sets of tRNA genes in these viral genomes accommodating AT-rich codons. These
additional tRNA gene sets enable these viruses to improve their overall translational effi ciency and
infect
Synechococcus
strains which have generally a high mol% G+C content (Enav
et al
., 2012).
The marine cyanophages described so far infect the coastal and oceanic
Synechococcus
strains.
In an attempt to fi nd out the cyanophages, from surface waters of Baltimore Inner Harbor and
Chesapeake Bay, infecting the estuarine strains of the same genus, Wang and Chen (2007) for the
fi rst time described one cyanomyovirus (S-CBM2 on host strain CB0101), three cyanosiphoviruses
(S-CBS2, S-CBS3 and S-CBS4 isolated on host strains CB0204, CB0202 and CB0101, respectively)