Biology Reference
In-Depth Information
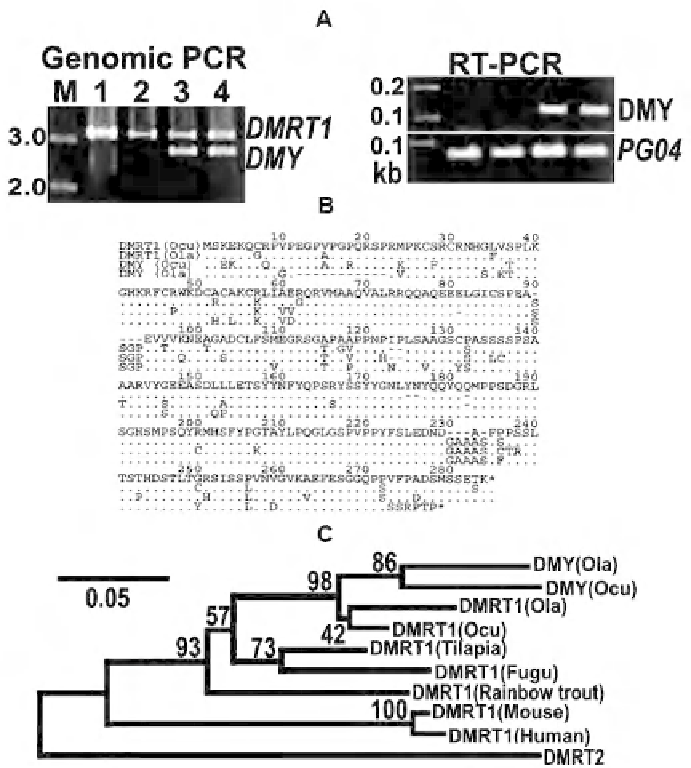
Fig. 9.
Upper panel (A): The
Dmrt1
and
Dmy
of
Oryzias latipes
are amplifi ed by the
Dmrt1
and
Dmy
of
O. curvinotus
Middle panel (B): Alignments of Dmy and Dmrt1 proteins of
O. latipes
(Ola) and
O. curvinotus
(Ocu) period indicated the sequence identity: dash a gap and asterix,
a stop codon. A single base pair deletion at position 272 of Ola
Dmy
has caused a frame-shift
mutation. Lower panel (C): A neighbor-joining tree based on the amino acid sequences from
positions 21 to 109 in the middle panel (from Matsuda et al., 2003; Zoological Science, 20:
159-161)
Herpin et al. (2010) have found that the
cis-
regulatory element containing
the
Dmrt1
binding site, pre-existing within
zanagi
element at the time of its
insertion, was co-opted to confer
Dmrt1bY
its expression pattern after gene
duplication. This fi nding reveals an important role for TEs in transcriptional
network rewiring and initiating a new function in the duplicated genes and
thereby creating new hierarchies of sex determining genes. Briefl y, despite