Biomedical Engineering Reference
In-Depth Information
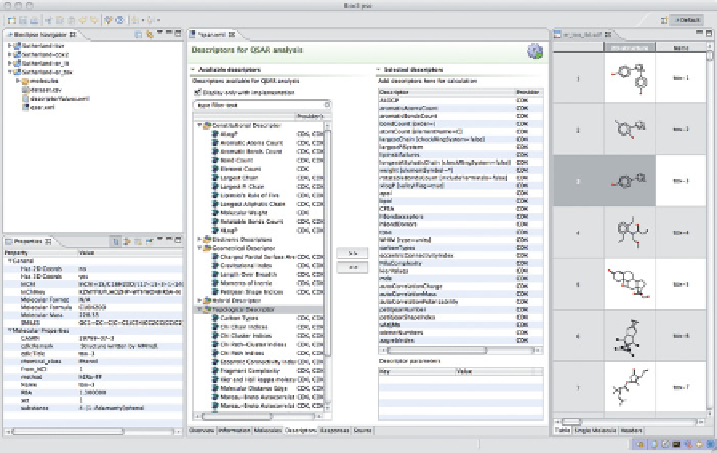
Figure 24.4
Screenshot showing creation of QSAR data set in Bioclipse, where a set
of molecules is aggregated and molecular descriptors are selected, creating a numerical
representation suitable for statistical modeling.
computer or via remote Web services. QSAR-ML also comes with a reference
implementation for the Bioclipse workbench [45, 46], which provides a graphi-
cal interface for setting up QSAR data sets, as shown in Figure 24.4. Prominent
features include adding and normalizing chemical structures in various formats,
cherry-picking local and remote descriptor implementations, adding responses
and metadata, and fi nally performing all calculations and exporting the com-
plete data set in QSAR-ML. Standardized QSAR opens up new ways to store,
query, and exchange analysis and makes it easy to join, extend, combine, and
work collectively with data.
24.3.2
Linking Knowledge Bases
One of the keys to collaboration is also the sharing of knowledge. A prominent
role here is in the linking of various databases, thereby allowing their integra-
tion and, where appropriate, their federation. Traditionally, linking databases
is done by using shared identifi ers. Well - known identifi ers for chemical struc-
tures include database - specifi c identifi ers such as the CAS registry number
[47], the PubChem compound identifi er [48], and the ChemSpider identifi er
[22] . When these are shared, they can be used to connect databases. Alternatively,
one could use an identifi er which can be calculated from the object itself. For
a wide set of small, organic molecules the InChI [49] fulfi lls this role [50].
An interesting proposal was made with the resource description framework
(RDF), [30] which suggests that a universal resource identifi er (URI) [51] is
Search WWH ::

Custom Search