Biomedical Engineering Reference
In-Depth Information
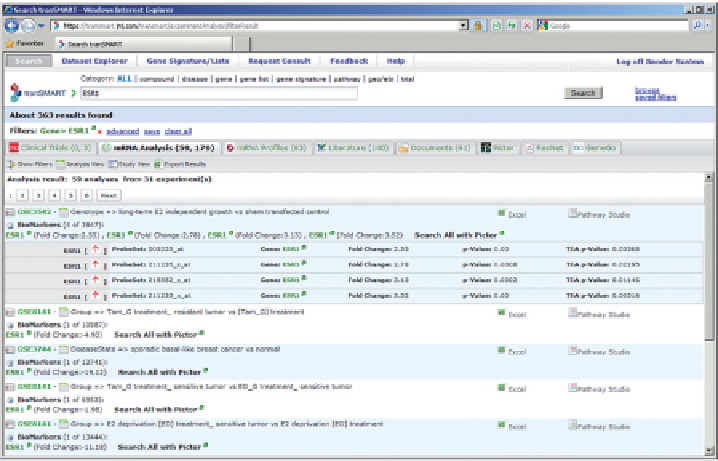
Figure 16.9
Simple search is illustrated by using a gene name (
ESR1
) as a search term.
The set of hits from different data sources are presented in a tabbed window. The
public gene expression hit list is shown.
carry out clinical trials in the disease of interest. Besides these dimensions one
should also consider if there is strong supporting scientifi c evidence for the
involvement of the targeted pathway in the pathophysiology of the disease.
We chose to establish this evidence statistically by mining a large corpus of
biological data such as gene expression experiments associated with diseases.
In order to accomplish that, we introduced the notion of signatures and con-
trasts in the data warehouse.
First, drug signatures can be uploaded—these are genes which are differ-
entially expressed when a drug candidate is added to an in vitro system which
is believed to mimic some aspects of the biology of a disease. Here the gene
expression is measured using a gene chip before applying the drug and after
and the difference is evaluated using, for example, a simple
t
- test. Genes
which pass quality control (QC) criteria and some predefi ned threshold then
comprise the signature of the drug candidate. These genes can be either
upregulated or downregulated and they are recorded as such in the
signature.
Second, we stored in the data warehouse a corpus of gene expression con-
trasts, that is, A versus B comparisons with associated diseases or drugs from
public databases described above. Each contrast contains a set of disregulated
genes (fold change with directionality and associated
p
- values) which pass
predefi ned quality criteria and metadata, including disease, drug, or phenotype
information. These contrasts are processed by Omicsoft Corporation using
Search WWH ::

Custom Search